6BWK
 
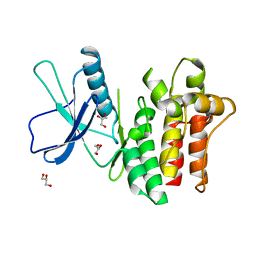 | |
1ZJE
 
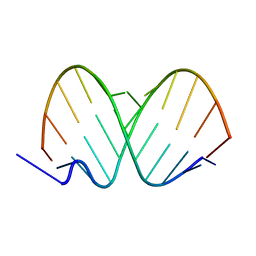 | | 12mer-spd | | Descriptor: | 5'-D(*AP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*T)-3', 5'-D(*TP*AP*GP*CP*CP*CP*CP*GP*CP*CP*CP*C)-3' | | Authors: | Dohm, J.A, Hsu, M.H, Hwu, J.R, Huang, R.C, Moudrianakis, E.N, Lattman, E.E, Gittis, A.G. | | Deposit date: | 2005-04-28 | | Release date: | 2005-05-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Influence of Ions, Hydration, and the Transcriptional Inhibitor P4N on the Conformations of the Sp1 Binding Site.
J.Mol.Biol., 349, 2005
|
|
4LF7
 
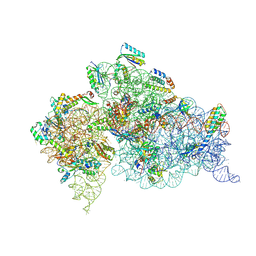 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, MAGNESIUM ION, PAROMOMYCIN, ... | | Authors: | Demirci, H, Belardinelli, R, Carr, J, Murphy IV, F, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-06-26 | | Release date: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (3.1484 Å) | | Cite: | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus
To be Published, 2013
|
|
1HWU
 
 | | STRUCTURE OF PII PROTEIN FROM HERBASPIRILLUM SEROPEDICAE | | Descriptor: | PII PROTEIN | | Authors: | Benelli, E.M, Buck, M, Polikarpov, I, De Souza, E.M, Cruz, L.M, Pedrosa, F.O. | | Deposit date: | 2001-01-10 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Herbaspirillum seropedicae signal transduction protein PII is structurally similar to the enteric GlnK.
Eur.J.Biochem., 269, 2002
|
|
4UZ1
 
 | |
1YYE
 
 | | Crystal structure of estrogen receptor beta complexed with way-202196 | | Descriptor: | 3-(3-FLUORO-4-HYDROXYPHENYL)-7-HYDROXY-1-NAPHTHONITRILE, Estrogen receptor beta, STEROID RECEPTOR COACTIVATOR-1 | | Authors: | Mewshaw, R.E, Edsall Jr, R.J, Yang, C, Manas, E.S, Xu, Z.B, Henderson, R.A, Keith Jr, J.C, Harris, H.A. | | Deposit date: | 2005-02-24 | | Release date: | 2006-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | ERbeta ligands. 3. Exploiting two binding orientations of the 2-phenylnaphthalene scaffold to achieve ERbeta selectivity
J.Med.Chem., 48, 2005
|
|
1SJY
 
 | | Crystal Structure of NUDIX HYDROLASE DR1025 FROM DEINOCOCCUS RADIODURANS | | Descriptor: | MutT/nudix family protein | | Authors: | Ranatunga, W, Hill, E.E, Mooster, J.L, Holbrook, E.L, Schulze-Gahmen, U, Xu, W, Bessman, M.J, Brenner, S.E, Holbrook, S.R, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-03-04 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural Studies of the Nudix Hydrolase DR1025 From Deinococcus radiodurans and its Ligand Complexes.
J.Mol.Biol., 339, 2004
|
|
6BR7
 
 | |
6JX0
 
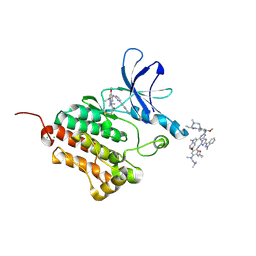 | |
1Q4K
 
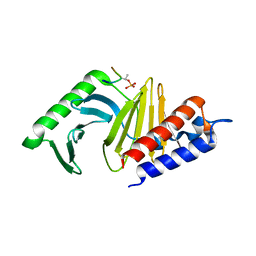 | | The polo-box domain of Plk1 in complex with a phospho-peptide | | Descriptor: | Phospho-peptide sequence Met.Gln.Ser.pThr.Pro.Leu, Serine/threonine-protein kinase PLK | | Authors: | Cheng, K, Lowe, E.D, Sinclair, J, Nigg, E.A, Johnson, L.N. | | Deposit date: | 2003-08-04 | | Release date: | 2003-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the human polo-like kinase-1 polo box domain and its phospho-peptide complex.
Embo J., 22, 2003
|
|
5AQN
 
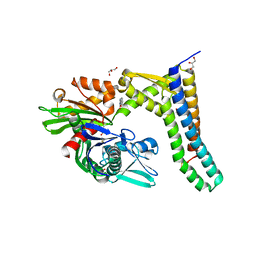 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, BENZOFURO[3,2-D]PYRIMIDIN-4(3H)-ONE, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
2HQ5
 
 | |
1YVQ
 
 | | The low salt (PEG) crystal structure of CO Hemoglobin E (betaE26K) approaching physiological pH (pH 7.5) | | Descriptor: | CARBON MONOXIDE, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Malashkevich, V.N, Balazs, T.C, Almo, S.C, Hirsch, R.E. | | Deposit date: | 2005-02-16 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of CO Hemoglobin E (betaE26K) approaching physiological pH (pH 7.5)
To be Published
|
|
5HVD
 
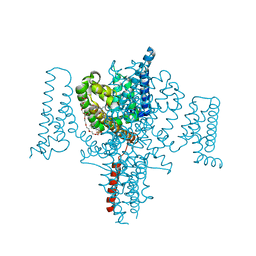 | | Full length Open-form Sodium Channel NavMs I218C | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DODECAETHYLENE GLYCOL, HEGA-10, ... | | Authors: | Sula, A, Booker, J.E, Naylor, C.E, Wallace, B.A. | | Deposit date: | 2016-01-28 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The complete structure of an activated open sodium channel.
Nat Commun, 8, 2017
|
|
2X8Y
 
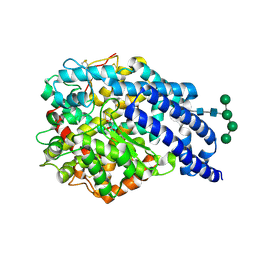 | | Crystal structure of AnCE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANGIOTENSIN CONVERTING ENZYME, CITRATE ANION, ... | | Authors: | Akif, M, Georgiadis, D, Mahajan, A, Dive, V, Sturrock, E.D, Isaac, R.E, Acharya, K.R. | | Deposit date: | 2010-03-14 | | Release date: | 2010-06-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High Resolution Crystal Structures of Drosophila Melanogaster Angiotensin Converting Enzyme in Complex with Novel Inhibitors and Anti- Hypertensive Drugs.
J.Mol.Biol., 400, 2010
|
|
6OI0
 
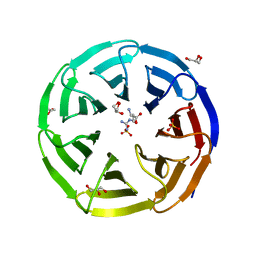 | | Crystal structure of human WDR5 in complex with L-arginine | | Descriptor: | ARGININE, GLYCEROL, SULFATE ION, ... | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-08 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
5A7U
 
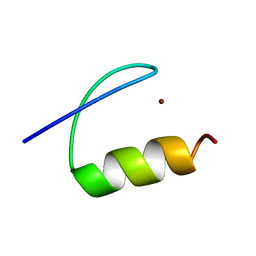 | | Single-particle cryo-EM of co-translational folded adr1 domain inside the E. coli ribosome exit tunnel. | | Descriptor: | REGULATORY PROTEIN ADR1, ZINC ION | | Authors: | Nilsson, O.B, Hedman, R, Marino, J, Wickles, S, Bischoff, L, Johansson, M, Muller-Lucks, A, Trovato, F, Puglisi, J.D, O'Brien, E, Beckmann, R, von Heijne, G. | | Deposit date: | 2015-07-10 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cotranslational Protein Folding Inside the Ribosome Exit Tunnel.
Cell Rep., 12, 2015
|
|
5KDH
 
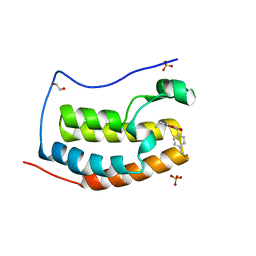 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH A DIHYDROPYRIDOPYRIMIDINE SCAFFOLD INHIBITOR | | Descriptor: | (5~{S})-1-ethyl-5-(4-methylphenyl)-8,9-dihydro-5~{H}-furo[3,4]pyrido[3,5-~{b}]pyrimidine-2,4,6-trione, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2016-06-08 | | Release date: | 2017-08-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | BET Bromodomain Inhibitors with One-Step Synthesis Discovered from Virtual Screen.
J. Med. Chem., 60, 2017
|
|
5KDU
 
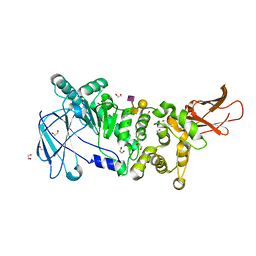 | | ZmpB metallopeptidase in complex with a2,6-Sialyl T-antigen | | Descriptor: | 1,2-ETHANEDIOL, F5/8 type C domain protein, SERINE, ... | | Authors: | Noach, I, Ficko-Blean, E, Stuart, C, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2X5N
 
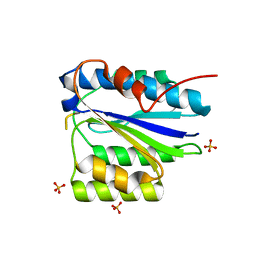 | | Crystal Structure of the SpRpn10 VWA domain | | Descriptor: | 26S PROTEASOME REGULATORY SUBUNIT RPN10, SULFATE ION | | Authors: | Riedinger, C, Boehringer, J, Trempe, J.-F, Lowe, E.D, Brown, N.R, Gehring, K, Noble, M.E.M, Gordon, C, Endicott, J.A. | | Deposit date: | 2010-02-10 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Structure of Rpn10 and its Interactions with Polyubiquitin Chains and the Proteasome Subunit Rpn12.
J.Biol.Chem., 285, 2010
|
|
8ASD
 
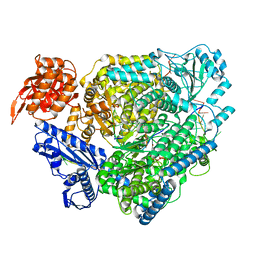 | | Structure of the SFTSV L protein stalled at late elongation [LATE-ELONGATION] | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, MAGNESIUM ION, RNA (5'-R(P*AP*AP*AP*AP*AP*AP*GP*AP*UP*CP*UP*GP*GP*GP*CP*GP*GP*UP*CP*UP*UP*UP*GP*UP*GP*U)-3'), ... | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Milewski, M, Busch, C, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2022-08-19 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into viral genome replication by the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 51, 2023
|
|
5A9K
 
 | | Structural basis for DNA strand separation by a hexameric replicative helicase | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, REPLICATION PROTEIN E1 | | Authors: | Chaban, Y, Stead, J.A, Ryzhenkova, K, Whelan, F, Lamber, K, Antson, F, Sanders, C.M, Orlova, E.V. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (19 Å) | | Cite: | Structural Basis for DNA Strand Separation by a Hexameric Replicative Helicase.
Nucleic Acids Res., 43, 2015
|
|
8AFR
 
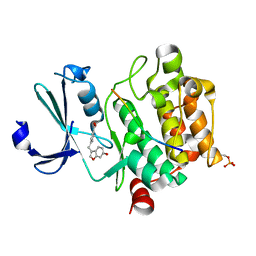 | | Pim1 in complex with 4-((6-hydroxybenzofuran-3-yl)methyl)benzoic acid and Pimtide | | Descriptor: | 4-((6-hydroxybenzofuran-3-yl)methyl)benzoic acid, Pimtide, Serine/threonine-protein kinase pim-1 | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2022-07-18 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
6D3I
 
 | | ftv7 dioxygenase with 2,4-D bound | | Descriptor: | (2,4-DICHLOROPHENOXY)ACETIC ACID, 2-OXOGLUTARIC ACID, COBALT (II) ION, ... | | Authors: | Rydel, T.J, Halls, C.E. | | Deposit date: | 2018-04-16 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.196 Å) | | Cite: | Development of enzymes for robust aryloxyphenoxypropionate and synthetic auxin herbicide tolerance traits in maize and soybean crops.
Pest Manag. Sci., 75, 2019
|
|
7SD5
 
 | | Crystallographic structure of neutralizing antibody 10-40 in complex with SARS-CoV-2 spike receptor binding domain | | Descriptor: | 10-40 Heavy chain, 10-40 Light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Reddem, E.R, Casner, R.G, Shapiro, L. | | Deposit date: | 2021-09-29 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | An antibody class with a common CDRH3 motif broadly neutralizes sarbecoviruses.
Sci Transl Med, 14, 2022
|
|
