8RTZ
 
 | | The structure of E. coli penicillin binding protein 3 (PBP3) in complex with a bicyclic peptide inhibitor | | Descriptor: | 1,1',1''-(1,3,5-triazinane-1,3,5-triyl)tripropan-1-one, Bicyclic peptide inhibitor, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Newman, H, Rowland, C.E, Dods, R, Lewis, N, Stanway, S.J, Bellini, D, Beswick, P. | | Deposit date: | 2024-01-29 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery and chemical optimisation of a Potent, Bi-cyclic (Bicycle) Antimicrobial Inhibitor of Escherichia coli PBP3
To Be Published
|
|
6PLP
 
 | | CryoEM structure of zebra fish alpha-1 glycine receptor YGF mutant bound with GABA in SMA, desensitized state | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PQT
 
 | |
7APD
 
 | | Bovine Papillomavirus E1 DNA helicase-replication fork complex | | Descriptor: | DNA (36-MER), DNA (40-MER), Replication protein E1 | | Authors: | Javed, A, Major, B, Stead, J, Sanders, C.M, Orlova, E.V. | | Deposit date: | 2020-10-16 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Unwinding of a DNA replication fork by a hexameric viral helicase.
Nat Commun, 12, 2021
|
|
6PSU
 
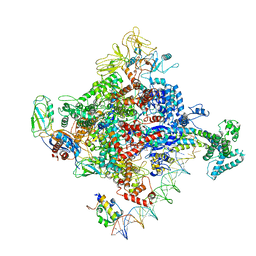 | | Escherichia coli RNA polymerase promoter unwinding intermediate (TRPi2) with TraR and rpsT P2 promoter | | Descriptor: | CHAPSO, DNA (85-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-07-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Stepwise Promoter Melting by Bacterial RNA Polymerase.
Mol.Cell, 78, 2020
|
|
8R1T
 
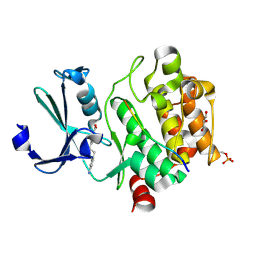 | | Pim1 in complex with 4-(4-aminophenethyl)benzoic acid and Pimtide | | Descriptor: | 4-(4-aminophenethyl)benzoic acid, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2023-11-02 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | What doesn't fit is made to fit: Pim-1 kinase adapts to the configuration of stilbene-based inhibitors.
Arch Pharm, 357, 2024
|
|
6YN4
 
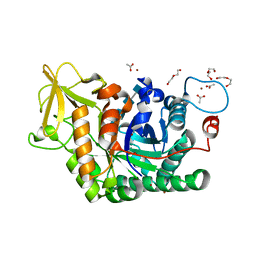 | | Structure of D169A/E171A double mutant of chitinase Chit42 from Trichoderma harzianum complexed with chitintetraose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2020-04-10 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural inspection and protein motions modelling of a fungal glycoside hydrolase family 18 chitinase by crystallography depicts a dynamic enzymatic mechanism
Comput Struct Biotechnol J, 19, 2021
|
|
8R1P
 
 | |
6Q0R
 
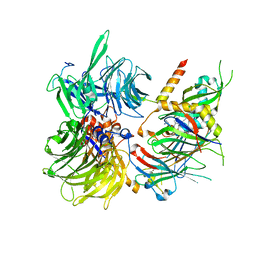 | | Structure of DDB1-DDA1-DCAF15 complex bound to E7820 and RBM39 | | Descriptor: | 3-cyano-N-(3-cyano-4-methyl-1H-indol-7-yl)benzene-1-sulfonamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Faust, T, Yoon, H, Nowak, R.P, Donovan, K.A, Li, Z, Cai, Q, Eleuteri, N.A, Zhang, T, Gray, N.S, Fischer, E.S. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15.
Nat.Chem.Biol., 16, 2020
|
|
6YLJ
 
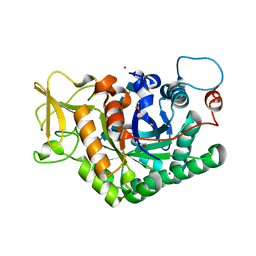 | | Structure of D169A/E171A double mutant of chitinase Chit42 from Trichoderma harzianum complexed with chitinhexaose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Endochitinase 42, ... | | Authors: | Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2020-04-07 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural inspection and protein motions modelling of a fungal glycoside hydrolase family 18 chitinase by crystallography depicts a dynamic enzymatic mechanism
Comput Struct Biotechnol J, 19, 2021
|
|
5N7X
 
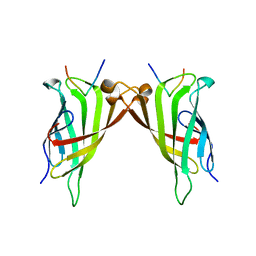 | | CRYSTAL STRUCTURE OF STREPTAVIDIN WITH PEPTIDE EWVHPQFEQKAK | | Descriptor: | GLU-TRP-VAL-HIS-PRO-GLN-PHE-GLU-GLN-LYS-ALA-LYS Peptide, Streptavidin | | Authors: | Lyamichev, V, Goodrich, L, Sullivan, E, Bannen, R, Benz, J, Albert, T, Patel, J. | | Deposit date: | 2017-02-21 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Stepwise Evolution Improves Identification of Diverse Peptides Binding to a Protein Target.
Sci Rep, 7, 2017
|
|
8S54
 
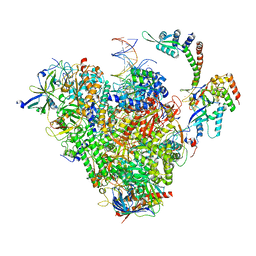 | | RNA polymerase II early elongation complex bound to TFIIE and TFIIF - state b (composite structure) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabber, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
5N8B
 
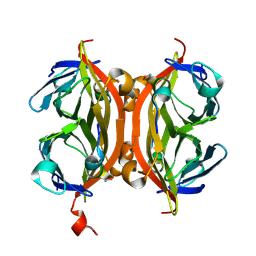 | | CRYSTAL STRUCTURE OF STREPTAVIDIN WITH PEPTIDE AFPDYLAEYHGG | | Descriptor: | ALA-PHE-PRO-ASP-TYR-LEU-ALA-GLU-TYR-HIS-GLY-GLY-NH2, Streptavidin | | Authors: | Lyamichev, V, Goodrich, L, Sullivan, E, Bannen, R, Benz, J, Albert, T, Patel, J. | | Deposit date: | 2017-02-23 | | Release date: | 2017-10-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Stepwise Evolution Improves Identification of Diverse Peptides Binding to a Protein Target.
Sci Rep, 7, 2017
|
|
8R1N
 
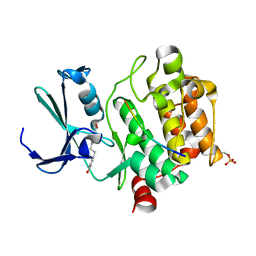 | |
6Q4B
 
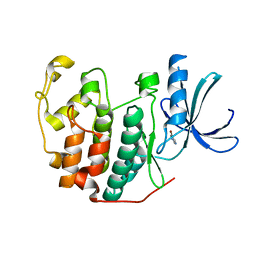 | | CDK2 in complex with FragLite13 | | Descriptor: | 5-bromanylpyrimidine, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
8BEO
 
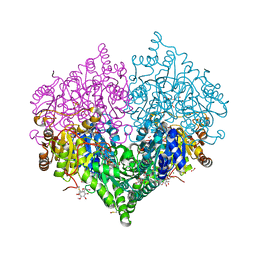 | | Crystal structure of E. coli glyoxylate carboligase mutant I393A with MAP | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2,3-DIMETHOXY-5-METHYL-1,4-BENZOQUINONE, ... | | Authors: | Shaanan, B, Binshtein, E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of E. coli glyoxylate carboligase mutant I393A with MAP
Not Published
|
|
6PS6
 
 | | XFEL beta2 AR structure by ligand exchange from Timolol to Timolol. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-1-(tert-butylamino)-3-[(4-morpholin-4-yl-1,2,5-thiadiazol-3-yl)oxy]propan-2-ol, CHOLESTEROL, ... | | Authors: | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
7B3X
 
 | | Notum complex with ARUK3003748 | | Descriptor: | 6-(m-tolylthio)-[1,2,4]triazolo[4,3-b]pyridazin-3(2H)-one, DIMETHYL SULFOXIDE, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Fish, P, Jones, E.Y. | | Deposit date: | 2020-12-01 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Virtual Screening Directly Identifies New Fragment-Sized Inhibitors of Carboxylesterase Notum with Nanomolar Activity.
J.Med.Chem., 65, 2022
|
|
6PSR
 
 | | Escherichia coli RNA polymerase promoter unwinding intermediate (TRPi1) with TraR and rpsT P2 promoter | | Descriptor: | CHAPSO, DNA (85-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-07-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Stepwise Promoter Melting by Bacterial RNA Polymerase.
Mol.Cell, 78, 2020
|
|
7B3I
 
 | | Notum complex with ARUK3003776 | | Descriptor: | 1,2-ETHANEDIOL, 3-(3-chlorophenyl)sulfanyl-1$l^{4},2,7,8-tetrazabicyclo[4.3.0]nona-1(6),2,4,7-tetraen-9-one, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2020-12-01 | | Release date: | 2021-12-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Virtual Screening Directly Identifies New Fragment-Sized Inhibitors of Carboxylesterase Notum with Nanomolar Activity.
J.Med.Chem., 65, 2022
|
|
8R10
 
 | |
7B3H
 
 | | Notum complex with ARUK3003909 | | Descriptor: | 1,2-ETHANEDIOL, 6-((3-(trifluoromethoxy)phenyl)thio)-[1,2,4]triazolo[4,3-b]pyridazin-3(2H)-one, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2020-11-30 | | Release date: | 2021-12-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Virtual Screening Directly Identifies New Fragment-Sized Inhibitors of Carboxylesterase Notum with Nanomolar Activity.
J.Med.Chem., 65, 2022
|
|
5NDC
 
 | | Structure of ba3-type cytochrome c oxidase from Thermus thermophilus by serial femtosecond crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide IIA, ... | | Authors: | Andersson, R, Safari, C, Dods, R, Nango, E, Tanaka, R, Nakane, T, Tono, K, Joti, Y, Bath, P, Dunevall, E, Bosman, E, Nureki, O, Iwata, S, Neutze, R, Branden, G. | | Deposit date: | 2017-03-08 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Serial femtosecond crystallography structure of cytochrome c oxidase at room temperature.
Sci Rep, 7, 2017
|
|
8R1L
 
 | | Structure of avian H5N1 influenza A polymerase in complex with human ANP32B. | | Descriptor: | Acidic leucine-rich nuclear phosphoprotein 32 family member B, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Carrique, L, Staller, E, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2023-11-02 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of H5N1 influenza polymerase with ANP32B reveal mechanisms of genome replication and host adaptation.
Nat Commun, 15, 2024
|
|
8W0X
 
 | |
