7EF0
 
 | |
7MBR
 
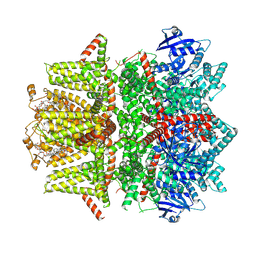 | | Cryo-EM structure of zebrafish TRPM5 in the presence of 6 uM calcium (apo state) | | 分子名称: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Ruan, Z, Lu, W, Du, J, Haley, E. | | 登録日 | 2021-04-01 | | 公開日 | 2021-07-07 | | 最終更新日 | 2021-07-28 | | 実験手法 | ELECTRON MICROSCOPY | | 主引用文献 | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MBS
 
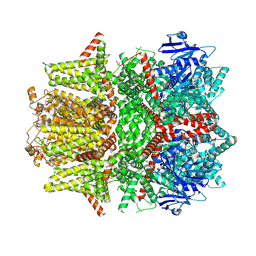 | | Cryo-EM structure of zebrafish TRPM5 in the presence of 6 uM calcium (open state) | | 分子名称: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Ruan, Z, Lu, W, Du, J, Haley, E. | | 登録日 | 2021-04-01 | | 公開日 | 2021-07-07 | | 最終更新日 | 2021-07-28 | | 実験手法 | ELECTRON MICROSCOPY | | 主引用文献 | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MBT
 
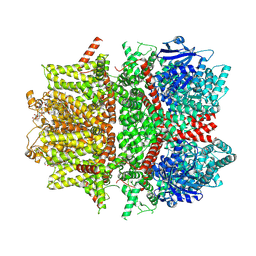 | | Cryo-EM structure of zebrafish TRPM5 E337A mutant in the presence of 5 mM calcium (low calcium occupancy in the transmembrane domain) | | 分子名称: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Ruan, Z, Lu, W, Du, J, Haley, E. | | 登録日 | 2021-04-01 | | 公開日 | 2021-07-07 | | 最終更新日 | 2021-07-28 | | 実験手法 | ELECTRON MICROSCOPY | | 主引用文献 | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MBQ
 
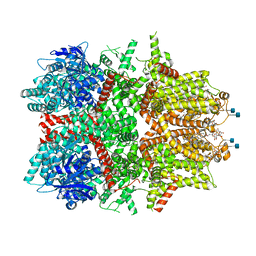 | | Cryo-EM structure of zebrafish TRPM5 in the presence of 5 mM calcium | | 分子名称: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Ruan, Z, Lu, W, Du, J, Haley, E. | | 登録日 | 2021-04-01 | | 公開日 | 2021-07-07 | | 最終更新日 | 2021-07-28 | | 実験手法 | ELECTRON MICROSCOPY (2.3 Å) | | 主引用文献 | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MBU
 
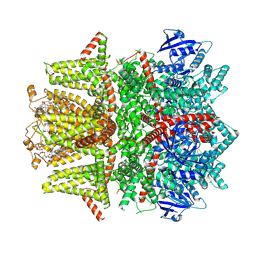 | | Cryo-EM structure of zebrafish TRPM5 E337A mutant in the presence of 5 mM calcium (high calcium occupancy in the transmembrane domain) | | 分子名称: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Ruan, Z, Lu, W, Du, J, Haley, E. | | 登録日 | 2021-04-01 | | 公開日 | 2021-07-07 | | 最終更新日 | 2021-07-28 | | 実験手法 | ELECTRON MICROSCOPY | | 主引用文献 | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MBV
 
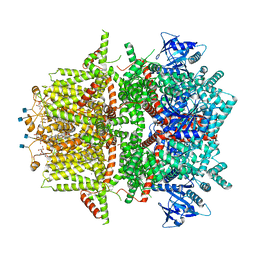 | | Cryo-EM structure of zebrafish TRPM5 in the presence of 5 mM calcium and 0.5 mM NDNA | | 分子名称: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Ruan, Z, Lu, W, Du, J, Haley, E. | | 登録日 | 2021-04-01 | | 公開日 | 2021-07-07 | | 最終更新日 | 2021-07-28 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MBP
 
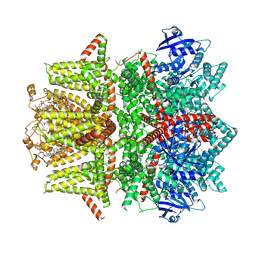 | | Cryo-EM structure of zebrafish TRPM5 in the presence of 1 mM EDTA | | 分子名称: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Ruan, Z, Lu, W, Du, J, Haley, E. | | 登録日 | 2021-04-01 | | 公開日 | 2021-09-22 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7YHP
 
 | |
7YHO
 
 | |
7YHQ
 
 | |
7W82
 
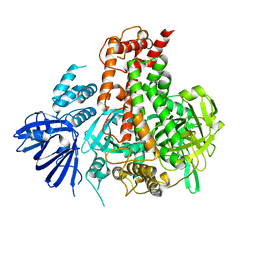 | | Crystal structure of maize RDR2 | | 分子名称: | RNA-dependent RNA polymerase | | 著者 | Du, X, Yang, Z, Du, J. | | 登録日 | 2021-12-07 | | 公開日 | 2022-06-08 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structure of plant RNA-DEPENDENT RNA POLYMERASE 2, an enzyme involved in small interfering RNA production.
Plant Cell, 34, 2022
|
|
7W84
 
 | |
7W88
 
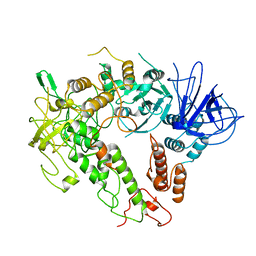 | |
7CCE
 
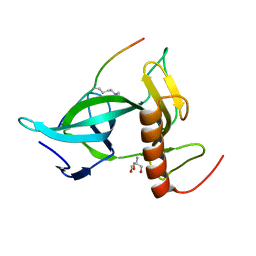 | |
6IP0
 
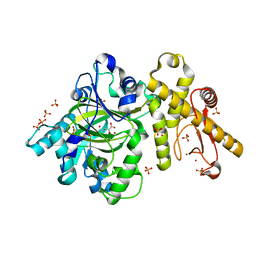 | |
6IP4
 
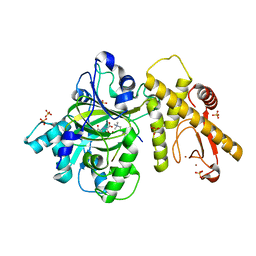 | |
6J9A
 
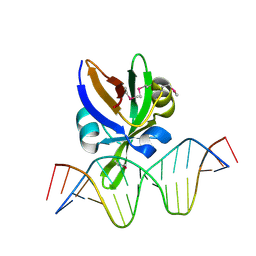 | | Crystal structure of Arabidopsis thaliana VAL1 in complex with FLC DNA fragment | | 分子名称: | B3 domain-containing transcription repressor VAL1, DNA (5'-D(*AP*AP*TP*CP*CP*AP*TP*GP*CP*AP*GP*AP*AP*TP*C)-3'), DNA (5'-D(*AP*TP*TP*CP*TP*GP*CP*AP*TP*GP*GP*AP*TP*TP*G)-3') | | 著者 | Hu, H, Du, J. | | 登録日 | 2019-01-22 | | 公開日 | 2019-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.915 Å) | | 主引用文献 | Embryonic resetting of the parental vernalized state by two B3 domain transcription factors in Arabidopsis.
Nat.Plants, 5, 2019
|
|
6J9C
 
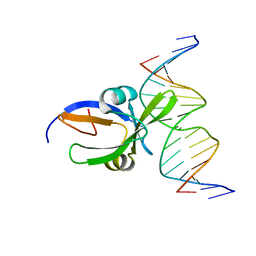 | | Crystal structure of Arabidopsis thaliana transcription factor LEC2-DNA complex | | 分子名称: | B3 domain-containing transcription factor LEC2, DNA (5'-D(*CP*AP*AP*TP*CP*CP*AP*TP*GP*CP*AP*GP*AP*AP*T)-3'), DNA (5'-D(*GP*AP*TP*TP*CP*TP*GP*CP*AP*TP*GP*GP*AP*TP*T)-3') | | 著者 | Hu, H, Du, J. | | 登録日 | 2019-01-22 | | 公開日 | 2019-05-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.102 Å) | | 主引用文献 | Embryonic resetting of the parental vernalized state by two B3 domain transcription factors in Arabidopsis.
Nat.Plants, 5, 2019
|
|
6J9B
 
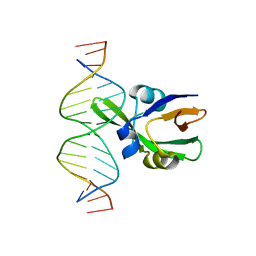 | | Arabidopsis FUS3-DNA complex | | 分子名称: | B3 domain-containing transcription factor FUS3, DNA (5'-D(*AP*AP*TP*CP*CP*AP*TP*GP*CP*AP*GP*AP*AP*TP*C)-3'), DNA (5'-D(*AP*TP*TP*CP*TP*GP*CP*AP*TP*GP*GP*AP*TP*TP*G)-3') | | 著者 | Hu, H, Du, J. | | 登録日 | 2019-01-22 | | 公開日 | 2019-05-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Embryonic resetting of the parental vernalized state by two B3 domain transcription factors in Arabidopsis.
Nat.Plants, 5, 2019
|
|
7VG2
 
 | |
7VG3
 
 | |
7VSP
 
 | |
7VSQ
 
 | |
8DGG
 
 | | Structure of glycosylated LAG-3 homodimer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Silberstein, J.L, Mathews, I.I, Frank, J.A, Chan, K.-W, Fernandez, D, Du, J, Wang, J, Kong, X.-P, Cochran, J.R. | | 登録日 | 2022-06-23 | | 公開日 | 2022-08-17 | | 最終更新日 | 2022-08-31 | | 実験手法 | X-RAY DIFFRACTION (3.78 Å) | | 主引用文献 | Structural basis for LAG-3 dimeric association and inhibition of T cell function
To Be Published
|
|
