3IU3
 
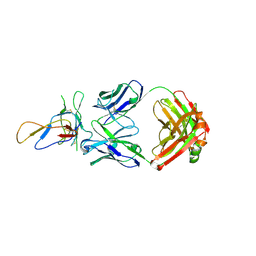 | | Crystal structure of the Fab fragment of therapeutic antibody Basiliximab in complex with IL-2Ra (CD25) ectodomain | | Descriptor: | Heavy chain of Fab fragment of Basiliximab, Interleukin-2 receptor alpha chain, Light chain of Fab fragment of Basiliximab, ... | | Authors: | Du, J, Yang, H, Wang, J, Ding, J. | | Deposit date: | 2009-08-29 | | Release date: | 2010-01-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the blockage of IL-2 signaling by therapeutic antibody basiliximab
J.Immunol., 184, 2010
|
|
3IWP
 
 | |
4EUE
 
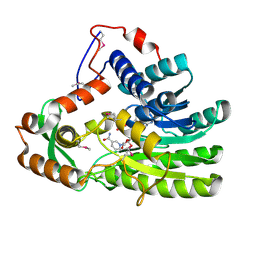 | | Crystal structure of Clostridium acetobutulicum trans-2-enoyl-CoA reductase in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Putative reductase CA_C0462, SODIUM ION | | Authors: | Hu, K, Zhao, M, Zhang, T, Yang, S, Ding, J. | | Deposit date: | 2012-04-25 | | Release date: | 2012-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of trans-2-enoyl-CoA reductases from Clostridium acetobutylicum and Treponema denticola: insights into the substrate specificity and the catalytic mechanism
Biochem.J., 449, 2013
|
|
3KT3
 
 | | Crystal structure of S. cerevisiae tryptophanyl-tRNA synthetase in complex with TrpAMP | | Descriptor: | SULFATE ION, TRYPTOPHANYL-5'AMP, Tryptophanyl-tRNA synthetase, ... | | Authors: | Zhou, M, Dong, X, Zhong, C, Shen, N, Ding, J. | | Deposit date: | 2009-11-24 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of Saccharomyces cerevisiae tryptophanyl-tRNA synthetase: new insights into the mechanism of tryptophan activation and implications for anti-fungal drug design
Nucleic Acids Res., 38, 2010
|
|
4EUH
 
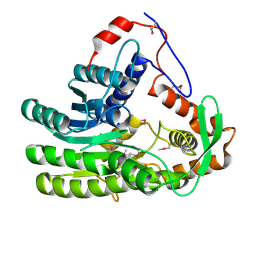 | | Crystal structure of Clostridium acetobutulicum trans-2-enoyl-CoA reductase apo form | | Descriptor: | Putative reductase CA_C0462, SODIUM ION | | Authors: | Hu, K, Zhao, M, Zhang, T, Yang, S, Ding, J. | | Deposit date: | 2012-04-25 | | Release date: | 2012-11-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of trans-2-enoyl-CoA reductases from Clostridium acetobutylicum and Treponema denticola: insights into the substrate specificity and the catalytic mechanism
Biochem.J., 449, 2013
|
|
3BM4
 
 | | Crystal Structure of Human ADP-ribose Pyrophosphatase NUDT5 In complex with magnesium and AMPcpr | | Descriptor: | ADP-sugar pyrophosphatase, ALPHA-BETA METHYLENE ADP-RIBOSE, MAGNESIUM ION | | Authors: | Zha, M, Guo, Q, Zhang, Y, Zhong, C, Ou, Y, Ding, J. | | Deposit date: | 2007-12-12 | | Release date: | 2008-05-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Mechanism of ADP-Ribose Hydrolysis By Human NUDT5 From Structural and Kinetic Studies
J.Mol.Biol., 379, 2008
|
|
3KT6
 
 | | Crystal structure of S. cerevisiae tryptophanyl-tRNA synthetase in complex with Trp | | Descriptor: | SULFATE ION, TRYPTOPHAN, Tryptophanyl-tRNA synthetase, ... | | Authors: | Zhou, M, Dong, X, Zhong, C, Shen, N, Ding, J. | | Deposit date: | 2009-11-24 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Saccharomyces cerevisiae tryptophanyl-tRNA synthetase: new insights into the mechanism of tryptophan activation and implications for anti-fungal drug design
Nucleic Acids Res., 38, 2010
|
|
3BKY
 
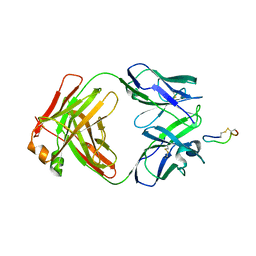 | | Crystal Structure of Chimeric Antibody C2H7 Fab in complex with a CD20 Peptide | | Descriptor: | B-lymphocyte antigen CD20, the Fab fragment of chimeric 2H7, heavy chain, ... | | Authors: | Du, J, Zhong, C, Ding, J. | | Deposit date: | 2007-12-07 | | Release date: | 2008-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of chimeric antibody C2H7 Fab in complex with a CD20 peptide
Mol.Immunol., 45, 2008
|
|
3KT8
 
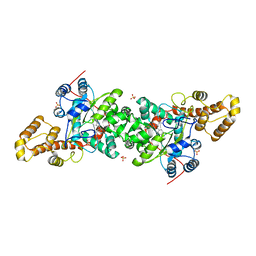 | | Crystal structure of S. cerevisiae tryptophanyl-tRNA synthetase in complex with L-tryptophanamide | | Descriptor: | L-TRYPTOPHANAMIDE, SULFATE ION, Tryptophanyl-tRNA synthetase, ... | | Authors: | Zhou, M, Dong, X, Zhong, C, Shen, N, Ding, J. | | Deposit date: | 2009-11-24 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Saccharomyces cerevisiae tryptophanyl-tRNA synthetase: new insights into the mechanism of tryptophan activation and implications for anti-fungal drug design
Nucleic Acids Res., 38, 2010
|
|
4EUF
 
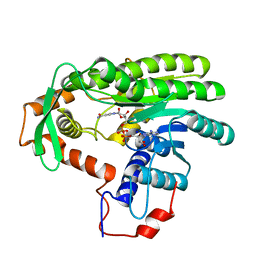 | | Crystal structure of Clostridium acetobutulicum trans-2-enoyl-CoA reductase in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative reductase CA_C0462, SODIUM ION | | Authors: | Hu, K, Zhao, M, Zhang, T, Yang, S, Ding, J. | | Deposit date: | 2012-04-25 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of trans-2-enoyl-CoA reductases from Clostridium acetobutylicum and Treponema denticola: insights into the substrate specificity and the catalytic mechanism
Biochem.J., 449, 2013
|
|
4FBG
 
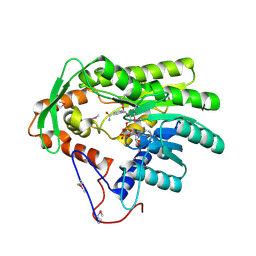 | | Crystal structure of Treponema denticola trans-2-enoyl-CoA reductase in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative reductase TDE_0597 | | Authors: | Hu, K, Zhao, M, Zhang, T, Yang, S, Ding, J. | | Deposit date: | 2012-05-23 | | Release date: | 2012-11-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structures of trans-2-enoyl-CoA reductases from Clostridium acetobutylicum and Treponema denticola: insights into the substrate specificity and the catalytic mechanism
Biochem.J., 449, 2013
|
|
4FGB
 
 | | Crystal structure of human calcium/calmodulin-dependent protein kinase I apo form | | Descriptor: | Calcium/calmodulin-dependent protein kinase type 1 | | Authors: | Zha, M, Zhong, C, Ou, Y, Wang, J, Han, L, Ding, J. | | Deposit date: | 2012-06-04 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of human CaMKIalpha reveal insights into the regulation mechanism of CaMKI.
Plos One, 7, 2012
|
|
4FG9
 
 | | Crystal structure of human calcium/calmodulin-dependent protein kinase I 1-320 in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Calcium/calmodulin-dependent protein kinase type 1 | | Authors: | Zha, M, Zhong, C, Ou, Y, Wang, J, Han, L, Ding, J. | | Deposit date: | 2012-06-04 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of human CaMKIalpha reveal insights into the regulation mechanism of CaMKI.
Plos One, 7, 2012
|
|
4FG8
 
 | | Crystal structure of human calcium/calmodulin-dependent protein kinase I 1-315 in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Calcium/calmodulin-dependent protein kinase type 1 | | Authors: | Zha, M, Zhong, C, Ou, Y, Wang, J, Han, L, Ding, J. | | Deposit date: | 2012-06-04 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of human CaMKIalpha reveal insights into the regulation mechanism of CaMKI.
Plos One, 7, 2012
|
|
6KMS
 
 | | Crystal structure of human N6amt1-Trm112 in complex with SAM (space group I422) | | Descriptor: | Methyltransferase N6AMT1, Multifunctional methyltransferase subunit TRM112-like protein, S-ADENOSYLMETHIONINE | | Authors: | Li, W.J, Shi, Y, Zhang, T.L, Ye, J, Ding, J.P. | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-18 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insight into human N6amt1-Trm112 complex functioning as a protein methyltransferase.
Cell Discov, 5, 2019
|
|
8HQN
 
 | | Activation mechanism of GPR132 by 9(S)-HODE | | Descriptor: | (9S,10E,12Z)-9-hydroxyoctadeca-10,12-dienoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
8HQM
 
 | | Activation mechanism of GPR132 by NPGLY | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
8HVI
 
 | | Activation mechanism of GPR132 by compound NOX-6-7 | | Descriptor: | 3-methyl-5-[(4-oxidanylidene-4-phenyl-butanoyl)amino]-1-benzofuran-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-26 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
8HQE
 
 | | Cryo-EM structure of the apo-GPR132-Gi | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
6KMR
 
 | | Crystal structure of human N6amt1-Trm112 in complex with SAM (space group P6122) | | Descriptor: | 1,2-ETHANEDIOL, Methyltransferase N6AMT1, Multifunctional methyltransferase subunit TRM112-like protein, ... | | Authors: | Li, W.J, Shi, Y, Zhang, T.L, Ye, J, Ding, J.P. | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into human N6amt1-Trm112 complex functioning as a protein methyltransferase.
Cell Discov, 5, 2019
|
|
3DEH
 
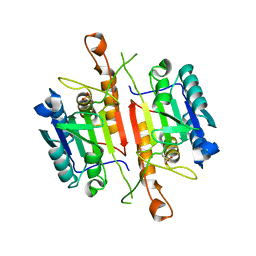 | | Crystal Structures of Caspase-3 with Bound Isoquinoline-1,3,4-trione Derivative Inhibitors | | Descriptor: | Caspase-3, isoquinoline-1,3,4(2H)-trione | | Authors: | Wu, J, Du, J, Li, J, Ding, J. | | Deposit date: | 2008-06-10 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Isoquinoline-1,3,4-trione Derivatives Inactivate Caspase-3 by Generation of Reactive Oxygen Species
J.Biol.Chem., 283, 2008
|
|
7X9E
 
 | | Crystal structure of the 76E1 Fab in complex with a SARS-CoV-2 spike peptide | | Descriptor: | 76E1 Fab Heavy Chain, 76E1 Fab Light Chain, Spike peptide | | Authors: | Chen, X, Zhang, T, Ding, J, Sun, X, Sun, B. | | Deposit date: | 2022-03-15 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Neutralization mechanism of a human antibody with pan-coronavirus reactivity including SARS-CoV-2.
Nat Microbiol, 7, 2022
|
|
3JXE
 
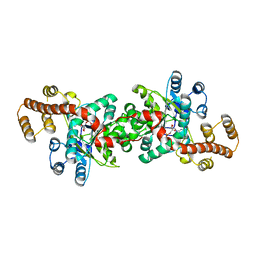 | | Crystal structure of Pyrococcus horikoshii tryptophanyl-tRNA synthetase in complex with TrpAMP | | Descriptor: | SULFATE ION, TRYPTOPHANYL-5'AMP, Tryptophanyl-tRNA synthetase | | Authors: | Zhou, M, Dong, X, Zhong, C, Shen, N, Yang, B, Ding, J. | | Deposit date: | 2009-09-19 | | Release date: | 2009-11-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of P. horikoshii tryptophanyl-tRNA synthetase and structure-based phylogenetic analysis suggest an archaeal origin of tryptophanyl-tRNA synthetase
To be Published
|
|
3DEJ
 
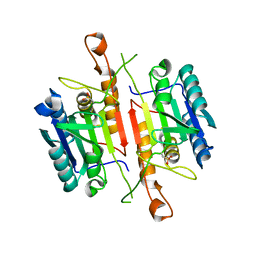 | | Crystal Structures of Caspase-3 with Bound Isoquinoline-1,3,4-trione Derivative Inhibitors | | Descriptor: | (1S)-1-(3-chlorophenyl)-2-oxo-2-[(1,3,4-trioxo-1,2,3,4-tetrahydroisoquinolin-5-yl)amino]ethyl acetate, Caspase-3 | | Authors: | Wu, J, Du, J, Li, J, Ding, J. | | Deposit date: | 2008-06-10 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Isoquinoline-1,3,4-trione Derivatives Inactivate Caspase-3 by Generation of Reactive Oxygen Species
J.Biol.Chem., 283, 2008
|
|
1XTS
 
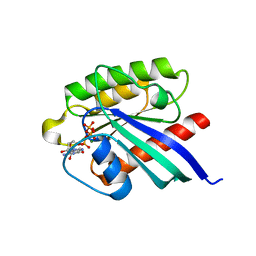 | | Structure of small GTPase human Rheb in complex with GTP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Yu, Y, Ding, J. | | Deposit date: | 2004-10-24 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Unique Biological Function of Small GTPase RHEB
J.Biol.Chem., 280, 2005
|
|
