8GNA
 
 | | Structure of the SbCas7-11-crRNA-NTR complex | | Descriptor: | RAMP superfamily protein, RNA (32-MER), RNA (5'-R(P*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*GP*GP*U)-3'), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-08-23 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Target RNA-guided protease activity in type III-E CRISPR-Cas system.
Nucleic Acids Res., 50, 2022
|
|
7WZB
 
 | | lipopolysaccharide assembly protein LapB (open) | | Descriptor: | Lipopolysaccharide assembly protein B, TRIETHYLENE GLYCOL, ZINC ION | | Authors: | Yan, L, Dong, H, Li, H, Liu, X, Deng, Z, Dong, C, Zhang, Z. | | Deposit date: | 2022-02-17 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | lipopolysaccharide assembly protein LapB (open)
To Be Published
|
|
8Z4L
 
 | | Cryo-EM structure of CTR-bound type VII CRISPR-Cas complex at substrate-engaged state I | | Descriptor: | RNA (40-MER), RNA (49-MER), ZINC ION, ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-04-17 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8Z4J
 
 | | Cryo-EM structure of CTR-bound type VII CRISPR-Cas complex at substrate-engaged state II | | Descriptor: | Protein structure, RNA (34-MER), RNA (38-MER), ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-04-17 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8Z99
 
 | | Cryo-EM structure of NTR-bound type VII CRISPR-Cas complex at substrate-engaged state +I | | Descriptor: | RNA (49-MER), RNA (54-MER), ZINC ION, ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
7XGL
 
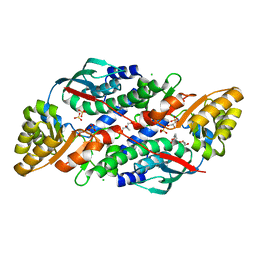 | | Quinolinate Phosphoribosyl Transferase (QAPRTase) from Streptomyces pyridomyceticus NRRL B-2517 in Apo form | | Descriptor: | CHLORIDE ION, GLYCEROL, Quinolinate Phosphoribosyl Transferase, ... | | Authors: | Zhou, Z, Yang, X, Huang, T, Wang, X, Liang, R, Zheng, J, Dai, S, Lin, S, Deng, Z. | | Deposit date: | 2022-04-05 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Bifunctional NadC Homologue PyrZ Catalyzes Nicotinic Acid Formation in Pyridomycin Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
7XGM
 
 | | Quinolinate Phosphoribosyl Transferase (QAPRTase) from Streptomyces pyridomyceticus NRRL B-2517 in complex with Quinolinic Acid (QA) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, QUINOLINIC ACID, ... | | Authors: | Zhou, Z, Yang, X, Huang, T, Wang, X, Liang, R, Zheng, J, Dai, S, Lin, S, Deng, Z. | | Deposit date: | 2022-04-05 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Bifunctional NadC Homologue PyrZ Catalyzes Nicotinic Acid Formation in Pyridomycin Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
7XGN
 
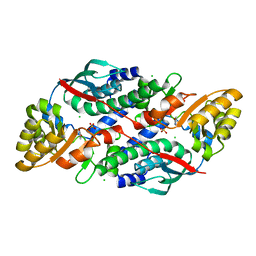 | | Quinolinate Phosphoribosyl Transferase (QAPRTase) from Streptomyces pyridomyceticus NRRL B-2517 in complex with Nicotinic Acid (NA) | | Descriptor: | CHLORIDE ION, NICOTINIC ACID, Quinolinate Phosphoribosyl Transferase, ... | | Authors: | Zhou, Z, Yang, X, Huang, T, Wang, X, Liang, R, Zheng, J, Dai, S, Lin, S, Deng, Z. | | Deposit date: | 2022-04-05 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Bifunctional NadC Homologue PyrZ Catalyzes Nicotinic Acid Formation in Pyridomycin Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
8YHD
 
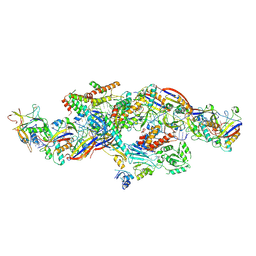 | | Cryo-EM structure of CTR-bound type VII CRISPR-Cas complex at post-state I | | Descriptor: | RNA (35-MER), RNA (53-MER), ZINC ION, ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-02-28 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8VUW
 
 | | ELIC5 with cysteamine in 2:1:1 POPC:POPE:POPG nanodisc in open conformation | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 2-AMINO-ETHANETHIOL, Erwinia chrysanthemi ligand-gated ion channel | | Authors: | Petroff II, J.T, Deng, Z, Rau, M.J, Fitzpatrick, J.A.J, Yuan, P, Cheng, W.W.L. | | Deposit date: | 2024-01-29 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Open-channel structure of a pentameric ligand-gated ion channel reveals a mechanism of leaflet-specific phospholipid modulation.
Nat Commun, 13, 2022
|
|
8HXB
 
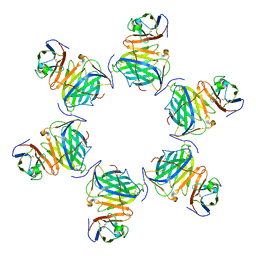 | | Cryo-EM structure of MPXV M2 hexamer in complex with human B7.2 | | Descriptor: | NFkB inhibitor, T-lymphocyte activation antigen CD86 | | Authors: | Wang, Y, Yang, S, Zhao, H, Deng, Z. | | Deposit date: | 2023-01-04 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural and functional insights into the modulation of T cell costimulation by monkeypox virus protein M2.
Nat Commun, 14, 2023
|
|
8HXC
 
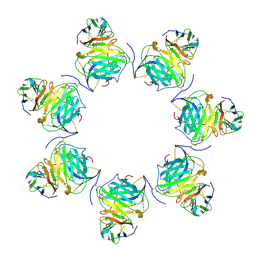 | | Cryo-EM structure of MPXV M2 heptamer in complex with human B7.2 | | Descriptor: | NFkB inhibitor, T-lymphocyte activation antigen CD86 | | Authors: | Wang, Y, Yang, S, Zhao, H, Deng, Z. | | Deposit date: | 2023-01-04 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural and functional insights into the modulation of T cell costimulation by monkeypox virus protein M2.
Nat Commun, 14, 2023
|
|
8HXA
 
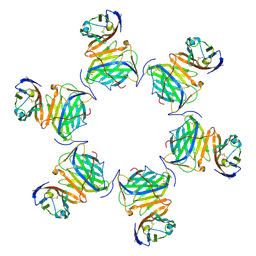 | | Cryo-EM structure of MPXV M2 in complex with human B7.1 | | Descriptor: | NFkB inhibitor, T-lymphocyte activation antigen CD80 | | Authors: | Wang, Y, Yang, S, Zhao, H, Deng, Z. | | Deposit date: | 2023-01-04 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural and functional insights into the modulation of T cell costimulation by monkeypox virus protein M2.
Nat Commun, 14, 2023
|
|
8XK8
 
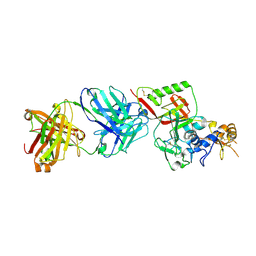 | | N1D10 Fab bound to SFTSV glycoprotein-Gn | | Descriptor: | Envelopment polyprotein, mAb N1D10 Fab heavy chain, mAb N1D10 Fab light chain | | Authors: | Zhao, H, Deng, Z. | | Deposit date: | 2023-12-22 | | Release date: | 2024-07-10 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | A broadly protective antibody targeting glycoprotein Gn inhibits severe fever with thrombocytopenia syndrome virus infection.
Nat Commun, 15, 2024
|
|
6CD2
 
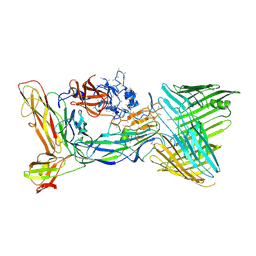 | | Crystal structure of the PapC usher bound to the chaperone-adhesin PapD-PapG | | Descriptor: | Chaperone protein PapD, Outer membrane usher protein PapC, PapGII adhesin protein | | Authors: | Omattage, N.S, Deng, Z, Yuan, P, Hultgren, S.J. | | Deposit date: | 2018-02-07 | | Release date: | 2018-10-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for usher activation and intramolecular subunit transfer in P pilus biogenesis in Escherichia coli.
Nat Microbiol, 3, 2018
|
|
8WWO
 
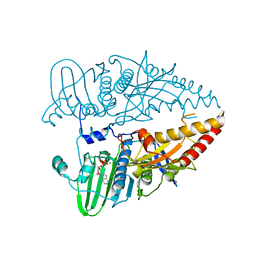 | |
8I87
 
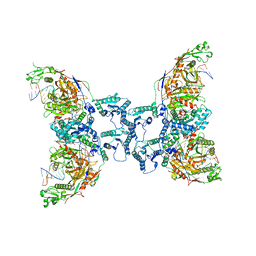 | | Cryo-EM structure of TIR-APAZ/Ago-gRNA-DNA complex | | Descriptor: | DNA (5'-D(P*TP*AP*TP*AP*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*A)-3'), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Zhang, H, Deng, Z.Q, Yu, G.M, Li, X.Z, Wang, X.S. | | Deposit date: | 2023-02-03 | | Release date: | 2023-07-19 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into mechanisms of Argonaute protein-associated NADase activation in bacterial immunity.
Cell Res., 33, 2023
|
|
8XAU
 
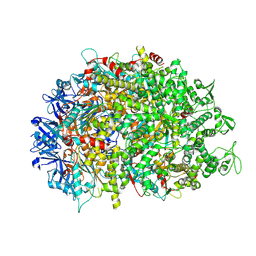 | | Cryo-EM structure of HerA | | Descriptor: | ATP-binding protein | | Authors: | Wang, Y, Deng, Z. | | Deposit date: | 2023-12-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Molecular and structural basis of an ATPase-nuclease dual-enzyme anti-phage defense complex.
Cell Res., 34, 2024
|
|
8XAV
 
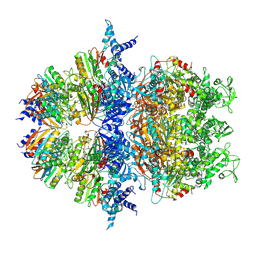 | | Cryo-EM structure of an anti-phage defense complex | | Descriptor: | ATP-binding protein, DUF4297 | | Authors: | Wang, Y, Deng, Z. | | Deposit date: | 2023-12-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Molecular and structural basis of an ATPase-nuclease dual-enzyme anti-phage defense complex.
Cell Res., 34, 2024
|
|
8XAW
 
 | |
8XAX
 
 | |
8XAY
 
 | |
8X7I
 
 | | Cryo-EM structures of RNF168/UbcH5c-Ub in complex with H2AK13Ub nucleosomes determined by intein-based E2-Ub-NCP conjugation strategy | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Ai, H.S, Tong, Z.B, Deng, Z.H, Pan, M, Liu, L. | | Deposit date: | 2023-11-24 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Capturing Snapshots of Nucleosomal H2A K13/K15 Ubiquitination Mediated by the Monomeric E3 Ligase RNF168
Biorxiv, 2024
|
|
8X7K
 
 | | Cryo-EM structures of RNF168/UbcH5c-Ub in complex with H2AK13Ub nucleosomes determined by activity-based chemical trapping strategy (adjacent H2AK13/15 dual-monoubiquitination) | | Descriptor: | DNA (143-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Ai, H.S, Tong, Z.B, Deng, Z.H, Pan, M, Liu, L. | | Deposit date: | 2023-11-24 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Capturing Snapshots of Nucleosomal H2A K13/K15 Ubiquitination Mediated by the Monomeric E3 Ligase RNF168
Biorxiv, 2024
|
|
8X7J
 
 | | Cryo-EM structures of RNF168/UbcH5c-Ub/nucleosomes complex determined by activity-based chemical trapping strategy | | Descriptor: | DNA (144-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Ai, H.S, Tong, Z.B, Deng, Z.H, Pan, M, Liu, L. | | Deposit date: | 2023-11-24 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Capturing Snapshots of Nucleosomal H2A K13/K15 Ubiquitination Mediated by the Monomeric E3 Ligase RNF168
Biorxiv, 2024
|
|
