1ZVB
 
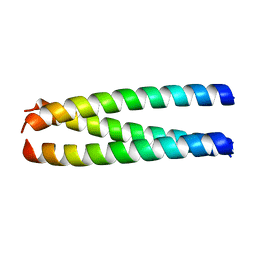 | | A structure-based mechanism of SARS virus membrane fusion | | Descriptor: | E2 glycoprotein | | Authors: | Deng, Y, Liu, J, Zheng, Q, Yong, W, Dai, J, Lu, M. | | Deposit date: | 2005-06-01 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures and Polymorphic Interactions of Two Heptad-Repeat Regions of the SARS Virus S2 Protein.
Structure, 14, 2006
|
|
1ZV8
 
 | | A structure-based mechanism of SARS virus membrane fusion | | Descriptor: | ACETATE ION, CACODYLATE ION, E2 glycoprotein, ... | | Authors: | Deng, Y, Liu, J, Zheng, Q, Yong, W, Dai, J, Lu, M. | | Deposit date: | 2005-06-01 | | Release date: | 2006-05-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures and Polymorphic Interactions of Two Heptad-Repeat Regions of the SARS Virus S2 Protein.
Structure, 14, 2006
|
|
1ZV7
 
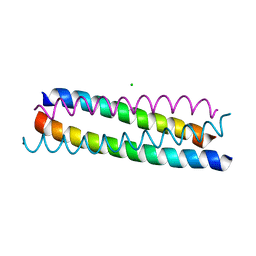 | | A structure-based mechanism of SARS virus membrane fusion | | Descriptor: | CHLORIDE ION, spike glycoprotein | | Authors: | Deng, Y, Liu, J, Zheng, Q, Yong, W, Dai, J, Lu, M. | | Deposit date: | 2005-06-01 | | Release date: | 2006-05-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures and Polymorphic Interactions of Two Heptad-Repeat Regions of the SARS Virus S2 Protein.
Structure, 14, 2006
|
|
1ZVA
 
 | | A structure-based mechanism of SARS virus membrane fusion | | Descriptor: | E2 glycoprotein | | Authors: | Deng, Y, Liu, J, Zheng, Q, Yong, W, Dai, J, Lu, M. | | Deposit date: | 2005-06-01 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures and Polymorphic Interactions of Two Heptad-Repeat Regions of the SARS Virus S2 Protein.
Structure, 14, 2006
|
|
2B1F
 
 | | Antiparallel four-stranded coiled coil specified by a 3-3-1 hydrophobic heptad repeat | | Descriptor: | General control protein GCN4 | | Authors: | Deng, Y, Liu, J, Zheng, Q, Eliezer, D, Kallenbach, N.R, Lu, M. | | Deposit date: | 2005-09-15 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Antiparallel four-stranded coiled coil specified by a 3-3-1 hydrophobic heptad repeat.
Structure, 14, 2006
|
|
2B22
 
 | | Antiparallel four-stranded coiled coil specified by a 3-3-1 hydrophobic heptad repeat | | Descriptor: | General control protein GCN4, SODIUM ION | | Authors: | Deng, Y, Liu, J, Zheng, Q, Eliezer, D, Kallenbach, N.R, Lu, M. | | Deposit date: | 2005-09-16 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Antiparallel four-stranded coiled coil specified by a 3-3-1 hydrophobic heptad repeat.
Structure, 14, 2006
|
|
4QYY
 
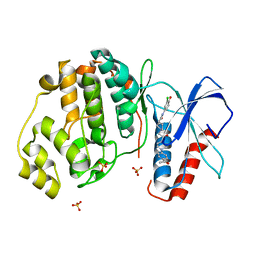 | | Discovery of Novel, Dual Mechanism ERK Inhibitors by Affinity Selection Screening of an Inactive Kinase State | | Descriptor: | (3R)-1-{2-[4-(4-acetylphenyl)piperazin-1-yl]-2-oxoethyl}-N-(3-chloro-4-hydroxyphenyl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Deng, Y, Shipps, G.W, Cooper, A, English, J.M, Annis, D.A, Carr, D, Nan, Y, Wang, T, Zhu, Y.H, Chuang, C, Dayananth, P, Hruza, A.W, Xiao, L, Jin, W, Kirschmeier, P, Windsor, W.T, Samatar, A.A. | | Deposit date: | 2014-07-26 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of Novel, Dual Mechanism ERK Inhibitors by Affinity Selection Screening of an Inactive Kinase.
J.Med.Chem., 57, 2014
|
|
6DAM
 
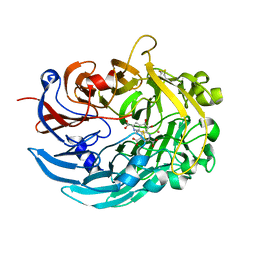 | | Crystal structure of lanthanide-dependent methanol dehydrogenase XoxF from Methylomicrobium buryatense 5G | | Descriptor: | LANTHANUM (III) ION, Lanthanide-dependent methanol dehydrogenase XoxF, PYRROLOQUINOLINE QUINONE, ... | | Authors: | Deng, Y, Ro, S.Y, Rosenzweig, A.C. | | Deposit date: | 2018-05-01 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and function of the lanthanide-dependent methanol dehydrogenase XoxF from the methanotroph Methylomicrobium buryatense 5GB1C.
J. Biol. Inorg. Chem., 23, 2018
|
|
7EN0
 
 | | Structure and Activity of SLAC1 Channels for Stomatal Signaling in Leaves | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, SLow Anion Channel 1, SPHINGOSINE | | Authors: | Deng, Y, Kashtoh, H, Wang, Q, Zhen, G, Li, Q, Tang, L, Gao, H, Zhang, C, Qin, L, Su, M, Li, F, Huang, X, Wang, Y, Xie, Q, Clarke, O.B, Hendrickson, W.A, Chen, Y. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure and activity of SLAC1 channels for stomatal signaling in leaves.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4UXL
 
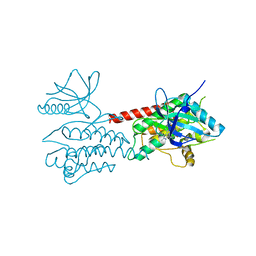 | | Structure of Human ROS1 Kinase Domain in Complex with PF-06463922 | | Descriptor: | (10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecine-3-carbonitrile, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE ROS | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A, Stewart, A. | | Deposit date: | 2014-08-25 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pf-06463922 is a Potent and Selective Next-Generation Ros1/Alk Inhibitor Capable of Blocking Crizotinib-Resistant Ros1 Mutations.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4W2R
 
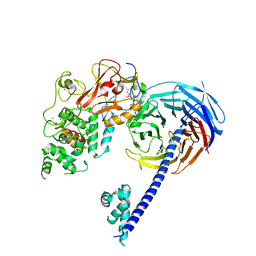 | | Structure of Hs/AcPRC2 in complex with 5,8-dichloro-2-[(4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-7-[(R)-methoxy(oxetan-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one | | Descriptor: | 5,8-dichloro-2-[(4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-7-[(R)-methoxy(oxetan-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one, Enhancer of zeste 2 polycomb repressive complex 2 subunit, Polycomb protein EED, ... | | Authors: | Gajiwala, K.S, Brooun, A, Liu, W, Deng, Y, Stewart, A.E. | | Deposit date: | 2017-09-25 | | Release date: | 2017-12-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Optimization of Orally Bioavailable Enhancer of Zeste Homolog 2 (EZH2) Inhibitors Using Ligand and Property-Based Design Strategies: Identification of Development Candidate (R)-5,8-Dichloro-7-(methoxy(oxetan-3-yl)methyl)-2-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-3,4-dihydroisoquinolin-1(2H)-one (PF-06821497).
J. Med. Chem., 61, 2018
|
|
5AAC
 
 | | Structure of C1156Y Mutant Human Anaplastic Lymphoma Kinase in Complex with Crizotinib | | Descriptor: | 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A, Stewart, A. | | Deposit date: | 2015-07-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Resensitization to Crizotinib by the Lorlatinib Alk Resistance Mutation L1198F.
N.Engl.J.Med., 374, 2016
|
|
6QYH
 
 | | Structure of Apo HPAB from E.coli | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-hydroxyphenylacetate 3-monooxygenase oxygenase component, CALCIUM ION, ... | | Authors: | Pecqueur, L, Lombard, M, Deng, Y, Fontecave, M. | | Deposit date: | 2019-03-09 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and Functional Characterization of 4-Hydroxyphenylacetate 3-Hydroxylase from Escherichia coli.
Chembiochem, 21, 2020
|
|
6QYI
 
 | | Structure of HPAB from E.coli in complex with FAD | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-hydroxyphenylacetate 3-monooxygenase oxygenase component, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pecqueur, L, Lombard, M, Deng, Y, Fontecave, M. | | Deposit date: | 2019-03-09 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Characterization of 4-Hydroxyphenylacetate 3-Hydroxylase from Escherichia coli.
Chembiochem, 21, 2020
|
|
4MOD
 
 | | Structure of the MERS-CoV fusion core | | Descriptor: | HR1 of S protein, LINKER, HR2 of S protein | | Authors: | Gao, J, Lu, G, Qi, J, Li, Y, Wu, Y, Deng, Y, Geng, H, Xiao, H, Tan, W, Yan, J, Gao, G.F. | | Deposit date: | 2013-09-12 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure of the fusion core and inhibition of fusion by a heptad repeat peptide derived from the S protein of Middle East respiratory syndrome coronavirus.
J.Virol., 87, 2013
|
|
2GUS
 
 | | Conformational Transition between Four- and Five-stranded Phenylalanine Zippers Determined by a Local Packing Interaction | | Descriptor: | Major outer membrane lipoprotein | | Authors: | Liu, J, Zheng, Q, Deng, Y, Kallenbach, N.R, Lu, M. | | Deposit date: | 2006-05-01 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Conformational Transition between Four and Five-stranded Phenylalanine Zippers Determined by a Local Packing Interaction.
J.Mol.Biol., 361, 2006
|
|
2HY6
 
 | | A seven-helix coiled coil | | Descriptor: | General control protein GCN4, HEXANE-1,6-DIOL | | Authors: | Liu, J, Zheng, Q, Deng, Y, Cheng, C.S, Kallenbach, N.R, Lu, M. | | Deposit date: | 2006-08-04 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A seven-helix coiled coil.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GUV
 
 | | Conformational Transition between Four- and Five-stranded Phenylalanine Zippers Determined by a Local Packing Interaction | | Descriptor: | Major outer membrane lipoprotein | | Authors: | Liu, J, Zheng, Q, Deng, Y, Kallenbach, N.R, Lu, M. | | Deposit date: | 2006-05-01 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational Transition between Four and Five-stranded Phenylalanine Zippers Determined by a Local Packing Interaction.
J.Mol.Biol., 361, 2006
|
|
2A7R
 
 | | Crystal structure of human Guanosine Monophosphate reductase 2 (GMPR2) | | Descriptor: | GMP reductase 2, GUANOSINE-5'-MONOPHOSPHATE, SULFATE ION | | Authors: | Li, J, Wei, Z, Zheng, M, Gu, X, Deng, Y, Qiu, R, Chen, F, Ji, C, Gong, W, Xie, Y, Mao, Y. | | Deposit date: | 2005-07-05 | | Release date: | 2006-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Human Guanosine Monophosphate Reductase 2 (GMPR2) in Complex with GMP
J.Mol.Biol., 355, 2006
|
|
1IA8
 
 | | THE 1.7 A CRYSTAL STRUCTURE OF HUMAN CELL CYCLE CHECKPOINT KINASE CHK1 | | Descriptor: | CHK1 CHECKPOINT KINASE, SULFATE ION | | Authors: | Chen, P, Luo, C, Deng, Y, Ryan, K, Register, J, Margosiak, S, Tempczyk-Russell, A, Nguyen, B, Myers, P, Lundgren, K, Chen Kan, C.-C, O'Connor, P.M. | | Deposit date: | 2001-03-22 | | Release date: | 2001-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.7 A crystal structure of human cell cycle checkpoint kinase Chk1: implications for Chk1 regulation.
Cell(Cambridge,Mass.), 100, 2000
|
|
8SIO
 
 | | Crystal structure of PRMT3 with YD1-66 | | Descriptor: | 5'-S-{3-[N'-(4'-chloro[1,1'-biphenyl]-3-yl)carbamimidamido]propyl}-5'-thioadenosine, Protein arginine N-methyltransferase 3 | | Authors: | Song, X, Dong, A, Arrowsmith, C.H, Edwards, A.M, Deng, Y, Huang, R, Min, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of PRMT3 with YD1-66
To be published
|
|
8SHR
 
 | | Crystal Structure of PRMT3 with Compound YD1-214 | | Descriptor: | 5'-S-[2-(phenylcarbamamido)ethyl]-5'-thioadenosine, Protein arginine N-methyltransferase 3 | | Authors: | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of PRMT3 with Compound YD1-214
To be published
|
|
8SHB
 
 | | Crystal Structure of PRMT3 with Compound YD1-208 | | Descriptor: | 5'-S-[3-(N'-phenylcarbamimidamido)propyl]-5'-thioadenosine, Protein arginine N-methyltransferase 3 | | Authors: | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-04-13 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of PRMT3 with Compound YD1-208
To be published
|
|
8SIH
 
 | | Crystal Structure of PRMT4 with Compound YD1-289 | | Descriptor: | 5'-{[2-(benzylcarbamamido)ethyl][3-(N'-cyclopentylcarbamimidamido)propyl]amino}-5'-deoxyadenosine, CALCIUM ION, Histone-arginine methyltransferase CARM1 | | Authors: | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of PRMT4 with Compound YD1-289
To be published
|
|
8SIG
 
 | | Crystal Structure of PRMT4 with Compound YD1-288 | | Descriptor: | 5'-{(3-aminopropyl)[2-(benzylcarbamamido)ethyl]amino}-5'-deoxyadenosine, Histone-arginine methyltransferase CARM1, SODIUM ION, ... | | Authors: | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of PRMT4 with Compound YD1-288
To be published
|
|
