1FVT
 
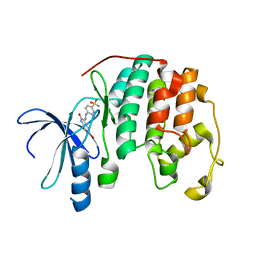 | | THE STRUCTURE OF CYCLIN-DEPENDENT KINASE 2 (CDK2) IN COMPLEX WITH AN OXINDOLE INHIBITOR | | Descriptor: | 4-[(2Z)-2-(5-bromo-2-oxo-1,2-dihydro-3H-indol-3-ylidene)hydrazinyl]benzene-1-sulfonamide, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Davis, S.T, Benson, B.G, Bramson, H.N, Chapman, D.E, Dickerson, S.H, Dold, K.M, Eberwein, D.J, Edelstein, M, Frye, S.V, Gampe Jr, R.T, Griffin, R.J, Harris, P.A, Hassell, A.M, Holmes, W.D, Hunter, R.N, Knick, V.B, Lackey, K, Lovejoy, B, Luzzio, M.J, Murray, D, Parker, P, Rocque, W.J, Shewchuk, L, Veal, J.M, Walker, D.H, Kuyper, L.K. | | Deposit date: | 2000-09-20 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Prevention of chemotherapy-induced alopecia in rats by CDK inhibitors.
Science, 291, 2001
|
|
1FVV
 
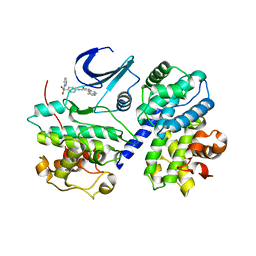 | | THE STRUCTURE OF CDK2/CYCLIN A IN COMPLEX WITH AN OXINDOLE INHIBITOR | | Descriptor: | 4-[(7-OXO-7H-THIAZOLO[5,4-E]INDOL-8-YLMETHYL)-AMINO]-N-PYRIDIN-2-YL-BENZENESULFONAMIDE, CYCLIN A, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Davis, S.T, Benson, B.G, Bramson, H.N, Chapman, D.E, Dickerson, S.H, Dold, K.M, Eberwein, D.J, Edelstein, M, Frye, S.V, Gampe Jr, R.T, Griffin, R.J, Harris, P.A, Hassell, A.M, Holmes, W.D, Hunter, R.N, Knick, V.B, Lackey, K, Lovejoy, B, Luzzio, M.J, Murray, D, Parker, P, Rocque, W.J, Shewchuk, L, Veal, J.M, Walker, D.H, Kuyper, L.K. | | Deposit date: | 2000-09-20 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Prevention of chemotherapy-induced alopecia in rats by CDK inhibitors.
Science, 291, 2001
|
|
6T9R
 
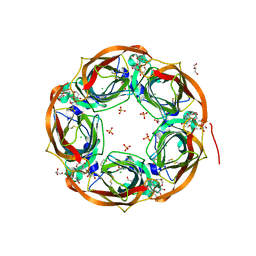 | | Aplysia californica AChBP in complex with a cytisine derivative | | Descriptor: | (1~{R},9~{S})-5-(3-oxidanylpropyl)-7,11-diazatricyclo[7.3.1.0^{2,7}]trideca-2,4-dien-6-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine binding protein, ... | | Authors: | Davis, S, Hunter, W.N. | | Deposit date: | 2019-10-28 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The thermodynamic profile and molecular interactions of a C(9)-cytisine derivative-binding acetylcholine-binding protein from Aplysia californica.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6QKK
 
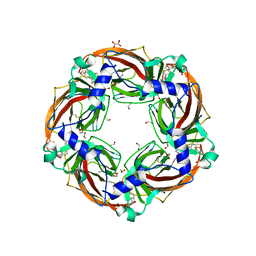 | | Aplysia californica AChBP in complex with 2-Fluoro-(carbamoylpyridinyl)deschloroepibatidine analogue (1) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[5-[(1~{R},2~{R},4~{S})-7-azabicyclo[2.2.1]heptan-2-yl]-2-fluoranyl-pyridin-3-yl]benzamide, ... | | Authors: | Davis, S, Bueno, R.V, Dawson, A, Hunter, W.N. | | Deposit date: | 2019-01-29 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interactions between 2'-fluoro-(carbamoylpyridinyl)deschloroepibatidine analogues and acetylcholine-binding protein inform on potent antagonist activity against nicotinic receptors
Acta Crystallogr.,Sect.D, 78, 2022
|
|
1BL0
 
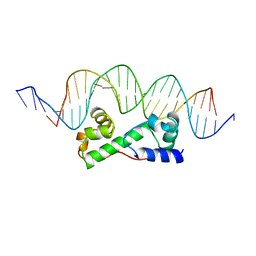 | | MULTIPLE ANTIBIOTIC RESISTANCE PROTEIN (MARA)/DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*TP*GP*CP*CP*AP*CP*GP*TP*TP*TP*TP*GP*CP*TP*AP*AP*AP*TP* CP*C)-3'), DNA (5'-D(*GP*GP*GP*GP*AP*TP*TP*TP*AP*GP*CP*AP*AP*AP*AP*CP*GP*TP*GP*GP*CP*AP* TP*C)-3'), PROTEIN (MULTIPLE ANTIBIOTIC RESISTANCE PROTEIN) | | Authors: | Davies, S, Rhee, R.G, Martin, J.L, Rosner, D.R. | | Deposit date: | 1998-07-22 | | Release date: | 1998-09-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1DFD
 
 | | OXIDISED DESULFOVIBRIO AFRICANUS FERREDOXIN I, NMR, 19 STRUCTURES | | Descriptor: | FERREDOXIN I, IRON/SULFUR CLUSTER | | Authors: | Davy, S.L, Osborne, M.J, Moore, G.R. | | Deposit date: | 1997-12-01 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Determination of the structure of oxidised Desulfovibrio africanus ferredoxin I by 1H NMR spectroscopy and comparison of its solution structure with its crystal structure.
J.Mol.Biol., 277, 1998
|
|
1DAX
 
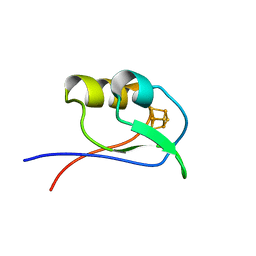 | | OXIDISED DESULFOVIBRIO AFRICANUS FERREDOXIN I, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | FERREDOXIN I, IRON/SULFUR CLUSTER | | Authors: | Davy, S.L, Osborne, M.J, Moore, G.R. | | Deposit date: | 1997-12-01 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Determination of the structure of oxidised Desulfovibrio africanus ferredoxin I by 1H NMR spectroscopy and comparison of its solution structure with its crystal structure.
J.Mol.Biol., 277, 1998
|
|
1HNF
 
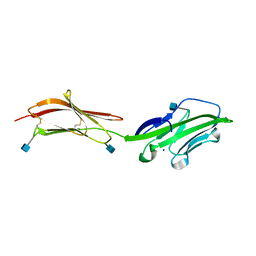 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR REGION OF THE HUMAN CELL ADHESION MOLECULE CD2 AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD2, SODIUM ION | | Authors: | Bodian, D.L, Jones, E.Y, Harlos, K, Stuart, D.I, Davis, S.J. | | Deposit date: | 1994-08-10 | | Release date: | 1995-02-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the extracellular region of the human cell adhesion molecule CD2 at 2.5 A resolution.
Structure, 2, 1994
|
|
2Z3R
 
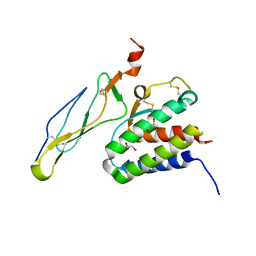 | | Crystal structure of the IL-15/IL-15Ra complex | | Descriptor: | GLYCEROL, Interleukin-15, Interleukin-15 receptor alpha chain | | Authors: | Chirifu, M, Yamagata, Y, Davis, S.J, Ikemizu, S. | | Deposit date: | 2007-06-05 | | Release date: | 2007-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the IL-15-IL-15Ralpha complex, a cytokine-receptor unit presented in trans
Nat.Immunol., 8, 2007
|
|
2Z3Q
 
 | | Crystal structure of the IL-15/IL-15Ra complex | | Descriptor: | Interleukin-15, Interleukin-15 receptor alpha chain | | Authors: | Chirifu, M, Yamagata, Y, Davis, S.J, Ikemizu, S. | | Deposit date: | 2007-06-05 | | Release date: | 2007-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the IL-15-IL-15Ralpha complex, a cytokine-receptor unit presented in trans
Nat.Immunol., 8, 2007
|
|
1HNG
 
 | | CRYSTAL STRUCTURE AT 2.8 ANGSTROMS RESOLUTION OF A SOLUBLE FORM OF THE CELL ADHESION MOLECULE CD2 | | Descriptor: | CD2 | | Authors: | Jones, E.Y, Davis, S.J, Williams, A.F, Harlos, K, Stuart, D.I. | | Deposit date: | 1994-08-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure at 2.8 A resolution of a soluble form of the cell adhesion molecule CD2.
Nature, 360, 1992
|
|
5FMV
 
 | | Crystal structure of human CD45 extracellular region, domains d1-d4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE C, SULFATE ION | | Authors: | Chang, V.T, Fernandes, R.A, Ganzinger, K.A, Lee, S.F, Siebold, C, McColl, J, Jonsson, P, Palayret, M, Harlos, K, Coles, C.H, Jones, E.Y, Lui, Y, Huang, E, Gilbert, R.J.C, Klenerman, D, Aricescu, A.R, Davis, S.J. | | Deposit date: | 2015-11-09 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Initiation of T Cell Signaling by Cd45 Segregation at 'Close Contacts'.
Nat.Immunol., 17, 2016
|
|
5FN6
 
 | | Crystal structure of human CD45 extracellular region, domains d1-d3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE C | | Authors: | Chang, V.T, Fernandes, R.A, Ganzinger, K.A, Lee, S.F, Siebold, C, McColl, J, Jonsson, P, Palayret, M, Harlos, K, Coles, C.H, Jones, E.Y, Lui, Y, Huang, E, Gilbert, R.J.C, Klenerman, D, Aricescu, A.R, Davis, S.J. | | Deposit date: | 2015-11-10 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Initiation of T Cell Signaling by Cd45 Segregation at 'Close Contacts'.
Nat.Immunol., 17, 2016
|
|
5FN8
 
 | | Crystal structure of rat CD45 extracellular region, domains d3-d4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRATE ANION, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE C | | Authors: | Chang, V.T, Fernandes, R.A, Ganzinger, K.A, Lee, S.F, Siebold, C, McColl, J, Jonsson, P, Palayret, M, Harlos, K, Coles, C.H, Jones, E.Y, Lui, Y, Huang, E, Gilbert, R.J.C, Klenerman, D, Aricescu, A.R, Davis, S.J. | | Deposit date: | 2015-11-11 | | Release date: | 2016-03-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Initiation of T Cell Signaling by Cd45 Segregation at 'Close Contacts'.
Nat.Immunol., 17, 2016
|
|
5FN7
 
 | | Crystal structure of human CD45 extracellular region, domains d1-d2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MERCURY (II) ION, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE C | | Authors: | Chang, V.T, Fernandes, R.A, Ganzinger, K.A, Lee, S.F, Siebold, C, McColl, J, Jonsson, P, Palayret, M, Harlos, K, Coles, C.H, Jones, E.Y, Lui, Y, Huang, E, Gilbert, R.J.C, Klenerman, D, Aricescu, A.R, Davis, S.J. | | Deposit date: | 2015-11-10 | | Release date: | 2016-03-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Initiation of T Cell Signaling by Cd45 Segregation at 'Close Contacts'.
Nat.Immunol., 17, 2016
|
|
3OSK
 
 | | Crystal structure of human CTLA-4 apo homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytotoxic T-lymphocyte protein 4, GLYCEROL | | Authors: | Yu, C, Sonnen, A.F.-P, Ikemizu, S, Stuart, D.I, Gilbert, R.J.C, Davis, S.J. | | Deposit date: | 2010-09-09 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rigid-body ligand recognition drives cytotoxic T-lymphocyte antigen 4 (CTLA-4) receptor triggering
J.Biol.Chem., 286, 2011
|
|
1ERH
 
 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE EXTRACELLULAR REGION OF THE COMPLEMENT REGULATORY PROTEIN, CD59, A NEW CELL SURFACE PROTEIN DOMAIN RELATED TO NEUROTOXINS | | Descriptor: | CD59 | | Authors: | Kieffer, B, Driscoll, P.C, Campbell, I.D, Willis, A.C, Van Der Merwe, P.A, Davis, S.J. | | Deposit date: | 1993-12-13 | | Release date: | 1994-04-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the extracellular region of the complement regulatory protein CD59, a new cell-surface protein domain related to snake venom neurotoxins.
Biochemistry, 33, 1994
|
|
1ERG
 
 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE EXTRACELLULAR REGION OF THE COMPLEMENT REGULATORY PROTEIN, CD59, A NEW CELL SURFACE PROTEIN DOMAIN RELATED TO NEUROTOXINS | | Descriptor: | CD59 | | Authors: | Kieffer, B, Driscoll, P.C, Campbell, I.D, Willis, A.C, Van Der Merwe, P.A, Davis, S.J. | | Deposit date: | 1993-12-13 | | Release date: | 1994-04-30 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the extracellular region of the complement regulatory protein CD59, a new cell-surface protein domain related to snake venom neurotoxins.
Biochemistry, 33, 1994
|
|
1R17
 
 | | Crystal Structure Analysis of S.epidermidis adhesin SdrG binding to Fibrinogen (adhesin-ligand complex) | | Descriptor: | CALCIUM ION, fibrinogen-binding protein SdrG, fibrinopeptide B | | Authors: | Ponnuraj, K, Bowden, M.G, Davis, S, Gurusiddappa, S, Moore, D, Choe, D, Xu, Y, Hook, M, Narayana, S.V.L. | | Deposit date: | 2003-09-23 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A "dock, lock and latch" Structural Model for a Staphylococcal Adhesin Binding to Fibrinogen
Cell(Cambridge,Mass.), 115, 2003
|
|
1R19
 
 | | Crystal Structure Analysis of S.epidermidis adhesin SdrG binding to Fibrinogen (Apo structure) | | Descriptor: | fibrinogen-binding protein SdrG | | Authors: | Ponnuraj, K, Bowden, M.G, Davis, S, Gurusiddappa, S, Moore, D, Choe, D, Xu, Y, Hook, M, Narayana, S.V.L. | | Deposit date: | 2003-09-23 | | Release date: | 2003-10-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | A "dock, lock and latch" Structural Model for a Staphylococcal Adhesin Binding to Fibrinogen
Cell(Cambridge,Mass.), 115, 2003
|
|
1NVQ
 
 | | The Complex Structure Of Checkpoint Kinase Chk1/UCN-01 | | Descriptor: | 7-HYDROXYSTAUROSPORINE, Peptide ASVSA, SULFATE ION, ... | | Authors: | Zhao, B, Bower, M.J, McDevitt, P.J, Zhao, H, Davis, S.T, Johanson, K.O, Green, S.M, Concha, N.O, Zhou, B.B. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Chk1 Inhibition by UCN-01
J.Biol.Chem., 277, 2002
|
|
1KE9
 
 | | CYCLIN-DEPENDENT KINASE 2 (CDK2) COMPLEXED WITH 3-{[4-({[AMINO(IMINO)METHYL]AMINOSULFONYL)ANILINO]METHYLENE}-2-OXO-2,3-DIHYDRO-1H-INDOLE | | Descriptor: | 3-{[4-([AMINO(IMINO)METHYL]AMINOSULFONYL)ANILINO]METHYLENE}-2-OXO-2,3-DIHYDRO-1H-INDOLE, Cell division protein kinase 2 | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.M, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
1KE7
 
 | | CYCLIN-DEPENDENT KINASE 2 (CDK2) COMPLEXED WITH 3-{[(2,2-DIOXIDO-1,3-DIHYDRO-2-BENZOTHIEN-5-YL)AMINO]METHYLENE}-5-(1,3-OXAZOL-5-YL)-1,3-DIHYDRO-2H-INDOL-2-ONE | | Descriptor: | 3-{[(2,2-DIOXIDO-1,3-DIHYDRO-2-BENZOTHIEN-5-YL)AMINO]METHYLENE}-5-(1,3-OXAZOL-5-YL)-1,3-DIHYDRO-2H-INDOL-2-ONE, Cell division protein kinase 2 | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.H, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
1KE5
 
 | | CDK2 complexed with N-methyl-4-{[(2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]amino}benzenesulfonamide | | Descriptor: | Cell division protein kinase 2, N-METHYL-4-{[(2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]AMINO}BENZENESULFONAMIDE | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.M, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
1NVS
 
 | | The Complex Structure Of Checkpoint Kinase Chk1/SB218078 | | Descriptor: | Peptide ASVSA, REL-(9R,12S)-9,10,11,12-TETRAHYDRO-9,12-EPOXY-1H-DIINDOLO[1,2,3-FG:3',2',1'-KL]PYRROLO[3,4-I][1,6]BENZODIAZOCINE-1,3(2H)-DIONE, SULFATE ION, ... | | Authors: | Zhao, B, Bower, M.J, McDevitt, P.J, Zhao, H, Davis, S.T, Johanson, K.O, Green, S.M, Concha, N.O, Zhou, B.B. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Chk1 Inhibition by UCN-01
J.Biol.Chem., 277, 2002
|
|
