1DWB
 
 | |
1DWD
 
 | |
1DWE
 
 | |
1DWC
 
 | |
3PR2
 
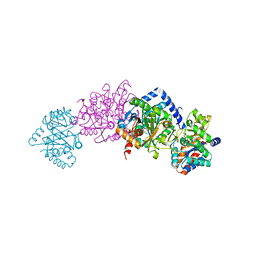 | | Tryptophan synthase indoline quinonoid structure with F9 inhibitor in alpha site | | Descriptor: | (Z)-N-[(1E)-1-carboxy-2-(2,3-dihydro-1H-indol-1-yl)ethylidene]{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methanaminium, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, CESIUM ION, ... | | Authors: | Lai, J, Niks, D, Wang, Y, Domratcheva, T, Barends, T.R.M, Schwarz, F, Olsen, R.A, Elliott, D.W, Fatmi, M.Q, Chang, C.A, Schlichting, I, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2010-11-29 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray and NMR Crystallography in an Enzyme Active Site: The Indoline Quinonoid Intermediate in Tryptophan Synthase.
J.Am.Chem.Soc., 133, 2011
|
|
3PYB
 
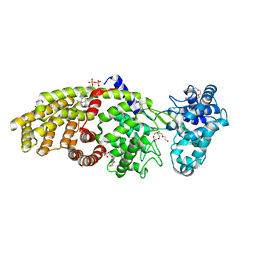 | | Crystal structure of ent-copalyl diphosphate synthase from Arabidopsis thaliana in complex with 13-aza-13,14-dihydrocopalyl diphosphate | | Descriptor: | 2-(methyl{2-[(1S,4aS,8aS)-5,5,8a-trimethyl-2-methylidenedecahydronaphthalen-1-yl]ethyl}amino)ethyl trihydrogen diphosphate, Ent-copalyl diphosphate synthase, chloroplastic, ... | | Authors: | Koksal, M, Christianson, D.W. | | Deposit date: | 2010-12-12 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.759 Å) | | Cite: | Structure of ent-Copalyl Diphosphate Synthase from Arabidopsis thaliana, a Protonation-Dependent Diterpene Cyclase
To be Published
|
|
3P1J
 
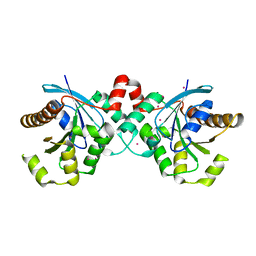 | | Crystal structure of human GTPase IMAP family member 2 in the nucleotide-free state | | Descriptor: | GTPase IMAP family member 2, UNKNOWN ATOM OR ION | | Authors: | Shen, L, Tempel, W, Tong, Y, Guan, X, Nedyalkova, L, Wernimont, A.K, Mackenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Andrews, D.W, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-30 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of human GTPase IMAP family member 2 in the nucleotide-free state
to be published
|
|
3P5R
 
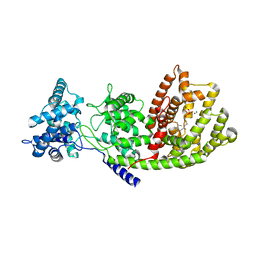 | | Crystal Structure of Taxadiene Synthase from Pacific Yew (Taxus brevifolia) in complex with Mg2+ and 2-fluorogeranylgeranyl diphosphate | | Descriptor: | (2Z,6E,10E)-2-fluoro-3,7,11,15-tetramethylhexadeca-2,6,10,14-tetraen-1-yl trihydrogen diphosphate, MAGNESIUM ION, Taxadiene synthase | | Authors: | Koksal, M, Christianson, D.W. | | Deposit date: | 2010-10-10 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | Taxadiene synthase structure and evolution of modular architecture in terpene biosynthesis.
Nature, 469, 2011
|
|
3OM2
 
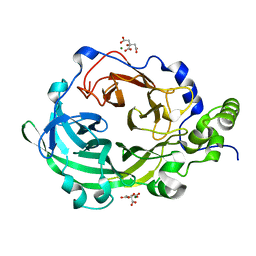 | | Crystal structure of B. megaterium levansucrase mutant D257A | | Descriptor: | CALCIUM ION, CITRIC ACID, Levansucrase, ... | | Authors: | Strube, C.P, Homann, A, Gamer, M, Jahn, D, Seibel, J, Heinz, D.W. | | Deposit date: | 2010-08-26 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Polysaccharide Synthesis of the Levansucrase SacB from Bacillus megaterium Is Controlled by Distinct Surface Motifs.
J.Biol.Chem., 286, 2011
|
|
104L
 
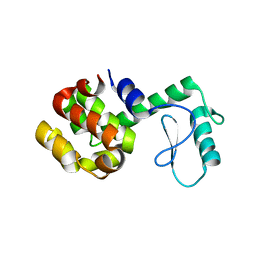 | |
103L
 
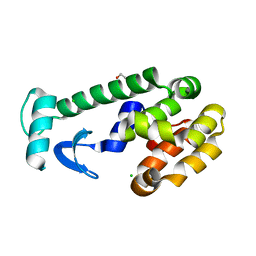 | |
1A42
 
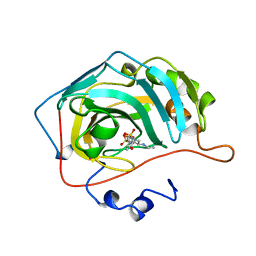 | | HUMAN CARBONIC ANHYDRASE II COMPLEXED WITH BRINZOLAMIDE | | Descriptor: | (4R)-2-(2-ethoxyethyl)-4-(ethylamino)-3,4-dihydro-2H-thieno[3,2-e][1,2]thiazine-6-sulfonamide 1,1-dioxide, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Boriack-Sjodin, P.A, Christianson, D.W. | | Deposit date: | 1998-02-10 | | Release date: | 1999-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of murine carbonic anhydrase IV and human carbonic anhydrase II complexed with brinzolamide: molecular basis of isozyme-drug discrimination.
Protein Sci., 7, 1998
|
|
1CR6
 
 | | CRYSTAL STRUCTURE OF MURINE SOLUBLE EPOXIDE HYDROLASE COMPLEXED WITH CPU INHIBITOR | | Descriptor: | EPOXIDE HYDROLASE, N-CYCLOHEXYL-N'-(PROPYL)PHENYL UREA | | Authors: | Argiriadi, M.A, Morisseau, C, Hammock, B.D, Christianson, D.W. | | Deposit date: | 1999-08-13 | | Release date: | 1999-11-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Detoxification of environmental mutagens and carcinogens: structure, mechanism, and evolution of liver epoxide hydrolase.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1CVC
 
 | |
1CIN
 
 | | THE POSITIONS OF HIS-64 AND A BOUND WATER IN HUMAN CARBONIC ANHYDRASE II UPON BINDING THREE STRUCTURALLY RELATED INHIBITORS | | Descriptor: | (4S-TRANS)-4-(METHYLAMINO)-5,6-DIHYDRO-6-METHYL-4H-THIENO(2,3-B)THIOPYRAN-2-SULFONAMIDE-7,7-DIOXIDE, CARBONIC ANHYDRASE II, METHYL MERCURY ION, ... | | Authors: | Smith, G.M, Alexander, R.S, Christianson, D.W, Mckeever, B.M, Ponticello, G.S, Springer, J.P, Randall, W.C, Baldwin, J.J, Habecker, C.N. | | Deposit date: | 1993-10-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Positions of His-64 and a bound water in human carbonic anhydrase II upon binding three structurally related inhibitors.
Protein Sci., 3, 1994
|
|
3Q9E
 
 | | Crystal structure of H159A APAH complexed with acetylspermine | | Descriptor: | Acetylpolyamine amidohydrolase, N-[3-({4-[(3-aminopropyl)amino]butyl}amino)propyl]acetamide, POTASSIUM ION, ... | | Authors: | Lombardi, P.M, Christianson, D.W. | | Deposit date: | 2011-01-07 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of prokaryotic polyamine deacetylase reveals evolutionary functional relationships with eukaryotic histone deacetylases .
Biochemistry, 50, 2011
|
|
3Q9B
 
 | | Crystal Structure of APAH complexed with M344 | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, Acetylpolyamine amidohydrolase, DIMETHYL SULFOXIDE, ... | | Authors: | Lombardi, P.M, Christianson, D.W. | | Deposit date: | 2011-01-07 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of prokaryotic polyamine deacetylase reveals evolutionary functional relationships with eukaryotic histone deacetylases .
Biochemistry, 50, 2011
|
|
1CNY
 
 | |
7PTD
 
 | | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C MUTANT R163K | | Descriptor: | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C | | Authors: | Heinz, D.W. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Probing the roles of active site residues in phosphatidylinositol-specific phospholipase C from Bacillus cereus by site-directed mutagenesis.
Biochemistry, 36, 1997
|
|
7RLS
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-68 | | Descriptor: | 3C-like proteinase, 6-[4-(3,4,5-trichlorophenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-26 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RNK
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-71 | | Descriptor: | 3C-like proteinase, 6-{4-[3-chloro-4-(hydroxymethyl)phenyl]piperazine-1-carbonyl}pyrimidine-2,4(3H,5H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-29 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RM2
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with Mcule-CSR-494190-S1 | | Descriptor: | 3C-like proteinase, 6-[4-(3,5-dichloro-4-methylphenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-26 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RME
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-52 | | Descriptor: | 3C-like proteinase, 6-{4-[4-chloro-3-(trifluoromethyl)phenyl]piperazine-1-carbonyl}pyrimidine-2,4(1H,3H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-27 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RMB
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-78 | | Descriptor: | 3C-like proteinase, 6-[4-(4-bromo-3-chlorophenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-27 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RN4
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-69 | | Descriptor: | 3C-like proteinase, 6-[4-(3,4-dichlorophenyl)piperidine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-29 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
