4RHK
 
 | |
4FXY
 
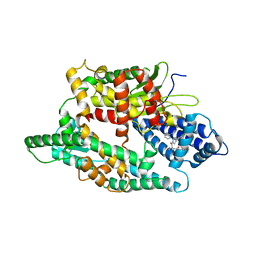 | | Crystal structure of rat neurolysin with bound pyrazolidin inhibitor | | 分子名称: | 1-{(2S)-1-[(3R)-3-(2-chlorophenyl)-2-(2-fluorophenyl)pyrazolidin-1-yl]-1-oxopropan-2-yl}-3-[(1R,3S,5R,7R)-tricyclo[3.3.1.1~3,7~]dec-2-yl]urea, Neurolysin, mitochondrial, ... | | 著者 | Rodgers, D.W, Hines, C.S. | | 登録日 | 2012-07-03 | | 公開日 | 2013-11-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Allosteric inhibition of the neuropeptidase neurolysin.
J.Biol.Chem., 289, 2014
|
|
4RHL
 
 | |
4RHQ
 
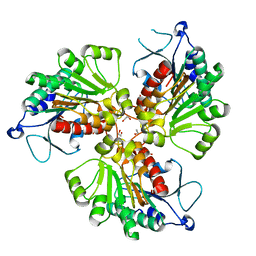 | | Crystal structure of T. brucei arginase-like protein double mutant S149D/S153D | | 分子名称: | 1,2-ETHANEDIOL, Arginase, GLYCEROL | | 著者 | Hai, Y, Barrett, M.P, Christianson, D.W. | | 登録日 | 2014-10-02 | | 公開日 | 2014-12-31 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Crystal Structure of an Arginase-like Protein from Trypanosoma brucei That Evolved without a Binuclear Manganese Cluster.
Biochemistry, 54, 2015
|
|
4RHJ
 
 | |
4RHM
 
 | | Crystal structure of T. brucei arginase-like protein quadruple mutant S149D/R151H/S153D/S226D | | 分子名称: | 1,2-ETHANEDIOL, Arginase, GLYCEROL, ... | | 著者 | Hai, Y, Barrett, M.P, Christianson, D.W. | | 登録日 | 2014-10-02 | | 公開日 | 2014-12-31 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal Structure of an Arginase-like Protein from Trypanosoma brucei That Evolved without a Binuclear Manganese Cluster.
Biochemistry, 54, 2015
|
|
6B15
 
 | | Crystal structure of CBMbc (family CBM26) from Eubacterium rectale Amy13K | | 分子名称: | 1,2-ETHANEDIOL, Amy13K | | 著者 | Cockburn, D.W, Wawrzak, Z, Perez Medina, K, Koropatkin, N.M. | | 登録日 | 2017-09-16 | | 公開日 | 2017-11-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Novel carbohydrate binding modules in the surface anchored alpha-amylase of Eubacterium rectale provide a molecular rationale for the range of starches used by this organism in the human gut.
Mol. Microbiol., 107, 2018
|
|
3CKE
 
 | | Crystal structure of aristolochene synthase in complex with 12,13-difluorofarnesyl diphosphate | | 分子名称: | (2E,6E)-12-fluoro-11-(fluoromethyl)-3,7-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, Aristolochene synthase, BETA-MERCAPTOETHANOL, ... | | 著者 | Shishova, E.Y, Yu, F, Miller, D.J, Faraldos, J.A, Zhao, Y, Coates, R.M, Allemann, R.K, Cane, D.E, Christianson, D.W. | | 登録日 | 2008-03-14 | | 公開日 | 2008-04-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | X-ray Crystallographic Studies of Substrate Binding to Aristolochene Synthase Suggest a Metal Ion Binding Sequence for Catalysis.
J.Biol.Chem., 283, 2008
|
|
4RN2
 
 | | Crystal structure of S39D HDAC8 in complex with a largazole analogue. | | 分子名称: | (5R,8S,11S)-5-methyl-8-(propan-2-yl)-11-[(1E)-4-sulfanylbut-1-en-1-yl]-3-thia-7,10,14,17,21-pentaazatricyclo[14.3.1.1~2,5~]henicosa-1(20),2(21),16,18-tetraene-6,9,13-trione, Histone deacetylase 8, POTASSIUM ION, ... | | 著者 | Decroos, C, Christianson, D.W. | | 登録日 | 2014-10-22 | | 公開日 | 2015-04-08 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Variable Active Site Loop Conformations Accommodate the Binding of Macrocyclic Largazole Analogues to HDAC8.
Biochemistry, 54, 2015
|
|
3CAA
 
 | | CLEAVED ANTICHYMOTRYPSIN A347R | | 分子名称: | ANTICHYMOTRYPSIN | | 著者 | Lukacs, C.M, Christianson, D.W. | | 登録日 | 1997-08-18 | | 公開日 | 1998-02-25 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Engineering an anion-binding cavity in antichymotrypsin modulates the "spring-loaded" serpin-protease interaction.
Biochemistry, 37, 1998
|
|
3HDS
 
 | | Crystal structure of 4-methylmuconolactone methylisomerase in complex with MES | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-methylmuconolactone methylisomerase, short peptide ASWSA | | 著者 | Marin, M, Heinz, D.W, Pieper, D.H, Klink, B.U. | | 登録日 | 2009-05-07 | | 公開日 | 2009-09-29 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Crystal structure and catalytic mechanism of 4-methylmuconolactone methylisomerase
J.Biol.Chem., 284, 2009
|
|
7TEH
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-2 | | 分子名称: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2022-01-05 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7TFR
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with NBH-2 | | 分子名称: | (1R,2S,5S)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-({1-[(2-methylpropane-2-sulfonyl)methyl]cyclohexyl}carbamoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2022-01-07 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7TDU
 
 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-1 | | 分子名称: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxo(1-~2~H)pyrrolidin-3-yl]propan-2-yl}-3-{N-[tert-butyl(~2~H)carbamoyl]-3-methyl-L-(N-~2~H)valyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-(~2~H)carboxamide, 3C-like proteinase | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2022-01-03 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | 主引用文献 | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
3G1R
 
 | | Crystal structure of human liver 5beta-reductase (AKR1D1) in complex with NADP and Finasteride. Resolution 1.70 A | | 分子名称: | (4aR,4bS,6aS,7S,9aS,9bS,11aR)-N-tert-butyl-4a,6a-dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxamide, 3-oxo-5-beta-steroid 4-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Di Costanzo, L, Drury, J.E, Penning, T.M, Christianson, D.W. | | 登録日 | 2009-01-30 | | 公開日 | 2009-06-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.701 Å) | | 主引用文献 | Inhibition of human steroid 5beta-reductase (AKR1D1) by finasteride and structure of the enzyme-inhibitor complex.
J.Biol.Chem., 284, 2009
|
|
4RZC
 
 | | Fv M6P-1 in complex with mannose-6-phosphate | | 分子名称: | 6-O-phosphono-alpha-D-mannopyranose, Fv M6P-1 heavy chain, Fv M6P-1 light chain, ... | | 著者 | Blackler, R.J, Evans, D.W, Evans, S.V, Muller-Loennies, S. | | 登録日 | 2014-12-19 | | 公開日 | 2015-11-04 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.723 Å) | | 主引用文献 | Single-chain antibody-fragment M6P-1 possesses a mannose 6-phosphate monosaccharide-specific binding pocket that distinguishes N-glycan phosphorylation in a branch-specific manner.
Glycobiology, 26, 2016
|
|
7T5H
 
 | | Structure of rabies virus phosphoprotein C-terminal domain, wild type | | 分子名称: | 1,2-ETHANEDIOL, PHOSPHATE ION, Phosphoprotein, ... | | 著者 | Zhan, J, Metcalfe, R.D, Gooley, P.R, Griffin, M.D.W. | | 登録日 | 2021-12-12 | | 公開日 | 2022-04-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Molecular Basis of Functional Effects of Phosphorylation of the C-Terminal Domain of the Rabies Virus P Protein.
J.Virol., 96, 2022
|
|
7T5G
 
 | | Structure of rabies virus phosphoprotein C-terminal domain, S210E mutant | | 分子名称: | Phosphoprotein, SULFATE ION | | 著者 | Zhan, J, Metcalfe, R.D, Gooley, P.R, Griffin, M.D.W. | | 登録日 | 2021-12-12 | | 公開日 | 2022-04-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Molecular Basis of Functional Effects of Phosphorylation of the C-Terminal Domain of the Rabies Virus P Protein.
J.Virol., 96, 2022
|
|
6CSS
 
 | |
3CPM
 
 | | plant peptide deformylase PDF1B crystal structure | | 分子名称: | Peptide deformylase, chloroplast, SULFATE ION, ... | | 著者 | Rodgers, D.W, Houtz, R.L, Dirk, L.M.A, Schmidt, J.J, Cai, Y. | | 登録日 | 2008-03-31 | | 公開日 | 2008-07-22 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Insights into the substrate specificity of plant peptide deformylase, an essential enzyme with potential for the development of novel biotechnology applications in agriculture
Biochem.J., 413, 2008
|
|
4E20
 
 | |
7SQ1
 
 | | BG505.MD39TS Env trimer in complex with Fab from antibody C05 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C05 Fab Light chain, ... | | 著者 | Moore, A, Du, J, Xu, Z, Walker, S, Kulp, D.W, Pallesen, J. | | 登録日 | 2021-11-04 | | 公開日 | 2022-06-22 | | 最終更新日 | 2022-12-07 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Induction of tier-2 neutralizing antibodies in mice with a DNA-encoded HIV envelope native like trimer.
Nat Commun, 13, 2022
|
|
4DPQ
 
 | | The structure of dihydrodipicolinate synthase 2 from Arabidopsis thaliana in complex with (S)-lysine | | 分子名称: | Dihydrodipicolinate synthase 2, chloroplastic, LYSINE, ... | | 著者 | Griffin, M.D.W, Billakanti, J.M, Gerrard, J.A, Dobson, R.C.J, Pearce, F.G. | | 登録日 | 2012-02-14 | | 公開日 | 2012-07-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.202 Å) | | 主引用文献 | Characterisation of the first enzymes committed to lysine biosynthesis in Arabidopsis thaliana
Plos One, 7, 2012
|
|
6CSP
 
 | |
3DD5
 
 | | Glomerella cingulata E600-cutinase complex | | 分子名称: | Cutinase, DIETHYL PHOSPHONATE | | 著者 | Nyon, M.P, Rice, D.W, Berrisford, J.M, Hounslow, A.M, Moir, A.J.G, Huang, H, Nathan, S, Mahadi, N.M, Farah Diba, A.B, Craven, C.J. | | 登録日 | 2008-06-05 | | 公開日 | 2008-11-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Catalysis by Glomerella cingulata Cutinase Requires Conformational Cycling between the Active and Inactive States of Its Catalytic Triad
J.Mol.Biol., 385, 2009
|
|
