5LRX
 
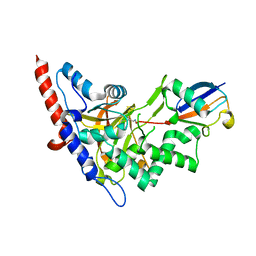 | | Structure of A20 OTU domain bound to ubiquitin | | Descriptor: | Polyubiquitin-B, Tumor necrosis factor alpha-induced protein 3 | | Authors: | Mevissen, T.E.T, Kulathu, Y, Mulder, M.P.C, Geurink, P.P, Maslen, S.L, Gersch, M, Elliott, P.R, Burke, J.E, van Tol, B.D.M, Akutsu, M, El Oualid, F, Kawasaki, M, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2016-08-22 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne.
Nature, 538, 2016
|
|
2YEY
 
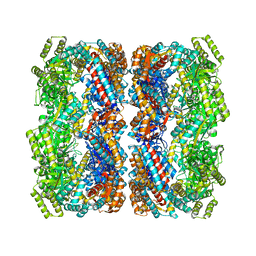 | | Crystal structure of the allosteric-defective chaperonin GroEL E434K mutant | | Descriptor: | 60 KDA CHAPERONIN | | Authors: | Cabo-Bilbao, A, Mechaly, A.E, Agirre, J, Spinelli, S, Sot, B, Muga, A, Guerin, D.M.A. | | Deposit date: | 2011-03-31 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Crystal Structure of the Temperature-Sensitive and Allosteric-Defective Chaperonin Groele461K.
J.Struct.Biol., 155, 2006
|
|
5NPR
 
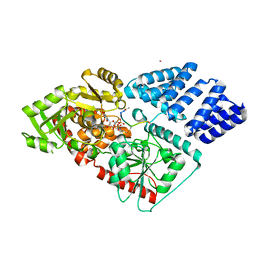 | | The human O-GlcNAc transferase in complex with a thiol-linked bisubstrate inhibitor | | Descriptor: | POTASSIUM ION, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] propyl hydrogen phosphate, ... | | Authors: | Rafie, K, van Aalten, D.M.F. | | Deposit date: | 2017-04-18 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Thio-Linked UDP-Peptide Conjugates as O-GlcNAc Transferase Inhibitors.
Bioconjug. Chem., 29, 2018
|
|
2WTK
 
 | |
5NDI
 
 | |
2X0Y
 
 | | Screening-based discovery of drug-like O-GlcNAcase inhibitor scaffolds | | Descriptor: | 7-[(2S)-2,3-DIHYDROXYPROPYL]-1,3-DIMETHYL-3,7-DIHYDRO-1H-PURINE-2,6-DIONE, O-GLCNACASE NAGJ | | Authors: | Dorfmueller, H.C, van Aalten, D.M.F. | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Screening-Based Discovery of Drug-Like O-Glcnacase Inhibitor Scaffolds
FEBS Lett., 584, 2010
|
|
5NEP
 
 | | The structure of the G. violaceus guanidine II riboswitch P1 stem-loop with methylguanidine | | Descriptor: | 1-METHYLGUANIDINE, RNA (5'-R(*GP*GP*UP*GP*GP*GP*GP*AP*CP*GP*AP*CP*CP*CP*CP*AP*(CBV)P*C)-3'), SODIUM ION, ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-03-11 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of the Guanidine-II Riboswitch.
Cell Chem Biol, 24, 2017
|
|
2XO0
 
 | | xpt-pbuX C74U Riboswitch from B. subtilis bound to 24-diamino-1,3,5- triazine identified by virtual screening | | Descriptor: | 1,3,5-TRIAZINE-2,4-DIAMINE, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Daldrop, P, Reyes, F.E, Robinson, D.A, Hammond, C.M, Lilley, D.M.J, Batey, R.T, Brenk, R. | | Deposit date: | 2010-08-09 | | Release date: | 2011-04-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel ligands for a purine riboswitch discovered by RNA-ligand docking.
Chem. Biol., 18, 2011
|
|
2XTK
 
 | |
352D
 
 | | THE CRYSTAL STRUCTURE OF A PARALLEL-STRANDED PARALLEL-STRANDED GUANINE TETRAPLEX AT 0.95 ANGSTROM RESOLUTION | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*GP*GP*GP*GP*T)-3'), SODIUM ION | | Authors: | Phillips, K, Dauter, Z, Murchie, A.I.H, Lilley, D.M.J, Luisi, B. | | Deposit date: | 1997-09-04 | | Release date: | 1997-11-10 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The crystal structure of a parallel-stranded guanine tetraplex at 0.95 A resolution.
J.Mol.Biol., 273, 1997
|
|
2XZ3
 
 | | BLV TM hairpin | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MALTOSE ABC TRANSPORTER PERIPLASMIC PROTEIN, ... | | Authors: | Schuettelkopf, A.W, Lamb, D, Brighty, D.W, van Aalten, D.M.F. | | Deposit date: | 2010-11-22 | | Release date: | 2011-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Charge-Surrounded Pockets and Electrostatic Interactions with Small Ions Modulate the Activity of Retroviral Fusion Proteins.
Plos Pathog., 7, 2011
|
|
5NZ3
 
 | | The structure of the thermobifida fusca guanidine III riboswitch with methylguanidine | | Descriptor: | 1-METHYLGUANIDINE, MAGNESIUM ION, RNA (41-MER), ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-05-12 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | Structure of the Guanidine III Riboswitch.
Cell Chem Biol, 24, 2017
|
|
2XNZ
 
 | | xpt-pbuX C74U Riboswitch from B. subtilis bound to acetoguanamine identified by virtual screening | | Descriptor: | 6-METHYL-1,3,5-TRIAZINE-2,4-DIAMINE, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Daldrop, P, Reyes, F.E, Robinson, D.A, Hammond, C.M, Lilley, D.M.J, Batey, R.T, Brenk, R. | | Deposit date: | 2010-08-06 | | Release date: | 2011-04-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Novel ligands for a purine riboswitch discovered by RNA-ligand docking.
Chem. Biol., 18, 2011
|
|
2XVP
 
 | |
5LRU
 
 | | Structure of Cezanne/OTUD7B OTU domain | | Descriptor: | OTU domain-containing protein 7B | | Authors: | Mevissen, T.E.T, Kulathu, Y, Mulder, M.P.C, Geurink, P.P, Maslen, S.L, Gersch, M, Elliott, P.R, Burke, J.E, van Tol, B.D.M, Akutsu, M, El Oualid, F, Kawasaki, M, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2016-08-22 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne.
Nature, 538, 2016
|
|
5NY8
 
 | | The structure of the thermobifida fusca guanidine III riboswitch with aminoguanidine | | Descriptor: | AMINOGUANIDINE, MAGNESIUM ION, RNA (41-MER), ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of the Guanidine III Riboswitch.
Cell Chem Biol, 24, 2017
|
|
5NZ6
 
 | |
2XNW
 
 | | XPT-PBUX C74U RIBOSWITCH FROM B. SUBTILIS BOUND TO A TRIAZOLO- TRIAZOLE-DIAMINE LIGAND IDENTIFIED BY VIRTUAL SCREENING | | Descriptor: | 3,6-diamino-1,5-dihydro[1,2,4]triazolo[4,3-b][1,2,4]triazol-4-ium, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Daldrop, P, Reyes, F.E, Robinson, D.A, Hammond, C.M, Lilley, D.M.J, Brenk, R. | | Deposit date: | 2010-08-06 | | Release date: | 2011-04-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel ligands for a purine riboswitch discovered by RNA-ligand docking.
Chem. Biol., 18, 2011
|
|
5NS3
 
 | |
5NZD
 
 | | The structure of the thermobifida fusca guanidine III riboswitch in space group P212121. | | Descriptor: | ACETATE ION, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-05-13 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structure of the Guanidine III Riboswitch.
Cell Chem Biol, 24, 2017
|
|
5NWQ
 
 | | The structure of the thermobifida fusca guanidine III riboswitch with guanidine. | | Descriptor: | GUANIDINE, Guanidine III riboswitch, MAGNESIUM ION, ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-05-08 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of the Guanidine III Riboswitch.
Cell Chem Biol, 24, 2017
|
|
2XO1
 
 | | xpt-pbuX C74U Riboswitch from B. subtilis bound to N6-methyladenine | | Descriptor: | ACETATE ION, COBALT HEXAMMINE(III), Guanine riboswitch, ... | | Authors: | Daldrop, P, Reyes, F.E, Robinson, D.A, Hammond, C.M, Lilley, D.M.J, Batey, R.T, Brenk, R. | | Deposit date: | 2010-08-09 | | Release date: | 2011-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Novel ligands for a purine riboswitch discovered by RNA-ligand docking.
Chem. Biol., 18, 2011
|
|
2XVN
 
 | | A. fumigatus chitinase A1 phenyl-methylguanylurea complex | | Descriptor: | 1-METHYL-3-(N-PHENYLCARBAMIMIDOYL)UREA, ASPERGILLUS FUMIGATUS CHITINASE A1, CHLORIDE ION | | Authors: | Rush, C.L, Schuttelkopf, A.W, Hurtado-Guerrero, R, Blair, D.E, Ibrahim, A.F.M, Desvergnes, S, Eggleston, I.M, van Aalten, D.M.F. | | Deposit date: | 2010-10-26 | | Release date: | 2010-11-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Natural Product-Guided Discovery of a Fungal Chitinase Inhibitor.
Chem.Biol., 17, 2010
|
|
5OAT
 
 | | PINK1 structure | | Descriptor: | MAGNESIUM ION, Serine/threonine-protein kinase PINK1, mitochondrial-like Protein | | Authors: | Kumar, A, Tamjar, J, Woodroof, H.I, Raimi, O.G, Waddell, A.Y, Peggie, M, Muqit, M.M.K, van Aalten, D.M.F. | | Deposit date: | 2017-06-23 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structure of PINK1 and mechanisms of Parkinson's disease associated mutations.
Elife, 6, 2017
|
|
5NS4
 
 | | Crystal structures of Cy3 cyanine fluorophores stacked onto the end of double-stranded RNA | | Descriptor: | 3-[(2~{Z})-2-[(~{E})-3-[3,3-dimethyl-1-(3-oxidanylpropyl)indol-1-ium-2-yl]prop-2-enylidene]-3,3-dimethyl-indol-1-yl]propan-1-ol, 50S ribosomal protein L5, MAGNESIUM ION, ... | | Authors: | Liu, Y.J, Lilley, D.M.J. | | Deposit date: | 2017-04-25 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Cyanine Fluorophores Stacked onto the End of Double-Stranded RNA.
Biophys. J., 113, 2017
|
|
