4CAU
 
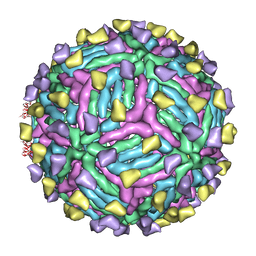 | | THREE-DIMENSIONAL STRUCTURE OF DENGUE VIRUS SEROTYPE 1 COMPLEXED WITH 2 HMAB 14C10 FAB | | Descriptor: | ENVELOPE PROTEIN E, FAB 14C10 | | Authors: | Teoh, E.P, Kukkaro, P, Teo, E.W, Lim, A.P, Tan, T.T, Yip, A, Schul, W, Aung, M, Kostyuchenko, V.A, Leo, Y.S, Chan, S.H, Smith, K.G, Chan, A.H, Zou, G, Ooi, E.E, Kemeny, D.M, Tan, G.K, Ng, J.K, Ng, M.L, Alonso, S, Fisher, D, Shi, P.Y, Hanson, B.J, Lok, S.M, Macary, P.A. | | Deposit date: | 2013-10-09 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | The Structural Basis for Serotype-Specific Neutralization of Dengue Virus by a Human Antibody.
Sci.Trans.Med, 4, 2012
|
|
4TVO
 
 | |
4C4M
 
 | | Crystal structure of the Sonic Hedgehog-chondroitin-4-sulphate complex | | Descriptor: | ACETATE ION, CALCIUM ION, SONIC HEDGEHOG PROTEIN, ... | | Authors: | Whalen, D.M, Malinauskas, T, Gilbert, R.J.C, Siebold, C. | | Deposit date: | 2013-09-05 | | Release date: | 2013-10-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural Insights Into Proteoglycan-Shaped Hedgehog Signaling.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4CN4
 
 | | GlgE isoform 1 from Streptomyces coelicolor E423A mutant with 2-deoxy- 2-fluoro-beta-maltosyl modification | | Descriptor: | ALPHA-1,4-GLUCAN:MALTOSE-1-PHOSPHATE MALTOSYLTRANSFERASE 1, alpha-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-beta-D-glucopyranose | | Authors: | Syson, K, Stevenson, C.E.M, Rashid, A.M, Saalbach, G, Tang, M, Tuukanen, A, Svergun, D.I, Withers, S.G, Lawson, D.M, Bornemann, S. | | Deposit date: | 2014-01-21 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insight Into How Streptomyces Coelicolor Maltosyl Transferase Glge Binds Alpha-Maltose 1-Phosphate and Forms a Maltosyl-Enzyme Intermediate.
Biochemistry, 53, 2014
|
|
4CN6
 
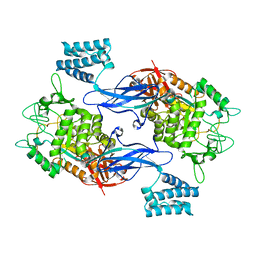 | | GlgE isoform 1 from Streptomyces coelicolor E423A mutant with maltose bound | | Descriptor: | ALPHA-1,4-GLUCAN:MALTOSE-1-PHOSPHATE MALTOSYLTRANSFERASE 1, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Syson, K, Stevenson, C.E.M, Rashid, A.M, Saalbach, G, Tang, M, Tuukanen, A, Svergun, D.I, Withers, S.G, Lawson, D.M, Bornemann, S. | | Deposit date: | 2014-01-21 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural Insight Into How Streptomyces Coelicolor Maltosyl Transferase Glge Binds Alpha-Maltose 1-Phosphate and Forms a Maltosyl-Enzyme Intermediate.
Biochemistry, 53, 2014
|
|
2RO5
 
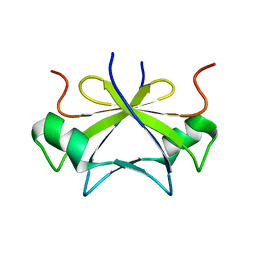 | | RDC-refined solution structure of the N-terminal DNA recognition domain of the Bacillus subtilis transition-state regulator SpoVT | | Descriptor: | Stage V sporulation protein T | | Authors: | Sullivan, D.M, Bobay, B.G, Kojetin, D.J, Thompson, R.J, Rance, M, Strauch, M.A, Cavanagh, J. | | Deposit date: | 2008-03-08 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Insights into the nature of DNA binding of AbrB-like transcription factors
Structure, 16, 2008
|
|
1OVN
 
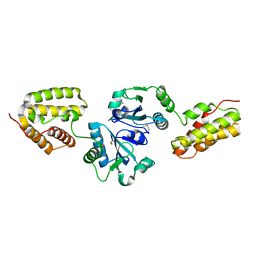 | | Crystal Structure and Functional Analysis of Drosophila Wind-- a PDI-Related Protein | | Descriptor: | CESIUM ION, Windbeutel | | Authors: | Ma, Q, Guo, C, Barnewitz, K, Sheldrick, G.M, Soling, H.D, Uson, I, Ferrari, D.M. | | Deposit date: | 2003-03-27 | | Release date: | 2004-02-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and functional analysis of Drosophila Wind, a protein-disulfide isomerase-related protein.
J.Biol.Chem., 278, 2003
|
|
2AQ2
 
 | | Crystal structure of T-cell receptor V beta domain variant complexed with superantigen SEC3 mutant | | Descriptor: | Enterotoxin type C-3, SODIUM ION, SULFATE ION, ... | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-17 | | Release date: | 2006-03-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
1P4F
 
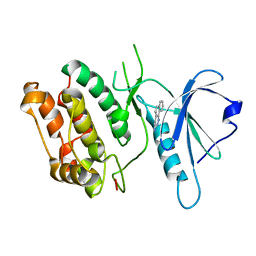 | | DEATH ASSOCIATED PROTEIN KINASE CATALYTIC DOMAIN WITH BOUND INHIBITOR FRAGMENT | | Descriptor: | 5,6-Dihydro-benzo[H]cinnolin-3-ylamine, Death-associated protein kinase 1 | | Authors: | Velentza, A.V, Wainwright, M.S, Zasadzki, M, Mirzoeva, S, Haiech, J, Focia, P.J, Egli, M, Watterson, D.M. | | Deposit date: | 2003-04-23 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An aminopyridazine-based inhibitor of a pro-apoptotic protein kinase attenuates hypoxia-ischemia induced acute brain injury.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1P7T
 
 | | Structure of Escherichia coli malate synthase G:pyruvate:acetyl-Coenzyme A abortive ternary complex at 1.95 angstrom resolution | | Descriptor: | ACETYL COENZYME *A, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Anstrom, D.M, Kallio, K, Remington, S.J. | | Deposit date: | 2003-05-05 | | Release date: | 2003-09-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the Escherichia Coli Malate Synthase G:pyruvate:acetyl-coenzyme A Abortive Ternary Complex at 1.95 Angstrom Resolution
Protein Sci., 12, 2003
|
|
1OIF
 
 | | Family 1 b-glucosidase from Thermotoga maritima | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, BETA-GLUCOSIDASE | | Authors: | Gloster, T, Zechel, D, Boraston, A.B, Boraston, C.M, Macdonald, J.M, Tilbrook, D.M, Stick, R.V, Davies, G.J. | | Deposit date: | 2003-06-16 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Iminosugar Glycosidase Inhibitors: Structural and Thermodynamic Dissection of the Binding of Isofagomine and 1-Deoxynojirimycin to Beta-Glucosidases
J.Am.Chem.Soc., 125, 2003
|
|
1OIM
 
 | | Family 1 b-glucosidase from Thermotoga maritima | | Descriptor: | 1-DEOXYNOJIRIMYCIN, BETA-GLUCOSIDASE A | | Authors: | Gloster, T, Zechel, D.L, Boraston, A.B, Boraston, C.M, Macdonald, J.M, Tilbrook, D.M, Stick, R.V, Davies, G.J. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Iminosugar Glycosidase Inhibitors: Structural and Thermodynamic Dissection of the Binding of Isofagomine and 1-Deoxynojirimycin to Beta-Glucosidases
J.Am.Chem.Soc., 125, 2003
|
|
2AQ1
 
 | | Crystal structure of T-cell receptor V beta domain variant complexed with superantigen SEC3 mutant | | Descriptor: | Enterotoxin type C-3, T-cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-17 | | Release date: | 2006-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2HEV
 
 | | Crystal structure of the complex between OX40L and OX40 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 4, Tumor necrosis factor receptor superfamily member 4 | | Authors: | Hymowitz, S.G, Compaan, D.M. | | Deposit date: | 2006-06-22 | | Release date: | 2006-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The Crystal Structure of the Costimulatory OX40-OX40L Complex.
Structure, 14, 2006
|
|
2HEW
 
 | | The X-ray crystal structure of murine OX40L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Tumor necrosis factor ligand superfamily member 4 | | Authors: | Hymowitz, S.G, Compaan, D.M. | | Deposit date: | 2006-06-22 | | Release date: | 2006-08-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Crystal Structure of the Costimulatory OX40-OX40L Complex.
Structure, 14, 2006
|
|
2UY8
 
 | | R92A mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | Authors: | Just, V.J, Burrell, M.R, Bowater, L, McRobbie, I, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-04-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Identity of the Active Site of Oxalate Decarboxylase and the Importance of the Stability of Active-Site Lid Conformations.
Biochem.J., 407, 2007
|
|
2UY9
 
 | | E162A mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | Authors: | Just, V.J, Burrell, M.R, Bowater, L, McRobbie, I, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-04-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Identity of the Active Site of Oxalate Decarboxylase and the Importance of the Stability of Active-Site Lid Conformations.
Biochem.J., 407, 2007
|
|
2DAA
 
 | |
2HQ3
 
 | | Solution NMR structure of the apo-NosL protein from Achromobacter cycloclastes | | Descriptor: | NosL protein | | Authors: | Taubner, L.M, McGuirl, M.A, Dooley, D.M, Copie, V. | | Deposit date: | 2006-07-18 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Studies of Apo Nosl, an Accessory Protein of the Nitrous Oxide Reductase System: Insights from Structural Homology with MerB, a Mercury Resistance Protein.
Biochemistry, 45, 2006
|
|
2HPU
 
 | | Solution NMR structure of the apo-NosL protein from Achromobacter cycloclastes | | Descriptor: | NosL protein | | Authors: | Taubner, L.M, McGuirl, M.A, Dooley, D.M, Copie, V. | | Deposit date: | 2006-07-17 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Studies of Apo Nosl, an Accessory Protein of the Nitrous Oxide Reductase System: Insights from Structural Homology with MerB, a Mercury Resistance Protein.
Biochemistry, 45, 2006
|
|
2IBK
 
 | | Bypass of Major Benzopyrene-dG Adduct by Y-Family DNA Polymerase with Unique Structural Gap | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 1,2-ETHANEDIOL, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*AP*T)-3', ... | | Authors: | Bauer, J, Ling, H, Sayer, J.M, Xing, G, Yagi, H, Jerina, D.M. | | Deposit date: | 2006-09-11 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A structural gap in Dpo4 supports mutagenic bypass of a major benzo[a]pyrene dG adduct in DNA through template misalignment.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3TVM
 
 | | Structure of the mouse CD1d-SMC124-iNKT TCR complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Girardi, E, Li, Y, Zajonc, D.M. | | Deposit date: | 2011-09-20 | | Release date: | 2012-02-01 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Glycolipids that Elicit IFN-gama-Biased Responses from Natural Killer T Cells
Chem.Biol., 18, 2011
|
|
9B9Z
 
 | | Structural mechanism of CB1R binding to peripheral and biased inverse agonists | | Descriptor: | (4S)-3-(4-chlorophenyl)-N'-[(1E)-ethanimidoyl]-4-phenyl-N-[4-(trifluoromethyl)benzene-1-sulfonyl]-4,5-dihydro-1H-pyrazole-1-carboximidamide, CNb36, Cannabinoid receptor 1,Glycogen synthase | | Authors: | Kumari, P, Dvoracsko, S, Enos, M.D, Ramesh, K, Lim, D, Hassan, S.A, Kunos, G, Cinar, R, Iyer, M.R, Rosenbaum, D.M. | | Deposit date: | 2024-04-03 | | Release date: | 2024-11-20 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural mechanism of CB 1 R binding to peripheral and biased inverse agonists.
Nat Commun, 15, 2024
|
|
9B9Y
 
 | | Structural mechanism of CB1R binding to peripheral and biased inverse agonists | | Descriptor: | CNb36, Cannabinoid receptor 1,Glycogen synthase, N-[(2S,3S)-4-(4-chlorophenyl)-3-(3-cyanophenyl)butan-2-yl]-2-methyl-2-{[5-(trifluoromethyl)pyridin-2-yl]oxy}propanamide | | Authors: | Kumari, P, Dvoracsko, S, Enos, M.D, Ramesh, K, Lim, D, Hassan, S.A, Kunos, G, Iyer, M.R, Rosenbaum, D.M. | | Deposit date: | 2024-04-03 | | Release date: | 2024-11-20 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural mechanism of CB 1 R binding to peripheral and biased inverse agonists.
Nat Commun, 15, 2024
|
|
2VVW
 
 | | Structure of Vaccinia virus protein A52 | | Descriptor: | PROTEIN A52 | | Authors: | Graham, S.C, Bahar, M.W, Cooray, S, Chen, R.A.-J, Whalen, D.M, Abrescia, N.G.A, Alderton, D, Owens, R.J, Stuart, D.I, Smith, G.L, Grimes, J.M. | | Deposit date: | 2008-06-12 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Vaccinia Virus Proteins A52 and B14 Share a Bcl-2-Like Fold But Have Evolved to Inhibit NF-kappaB Rather Than Apoptosis
Plos Pathog., 4, 2008
|
|
