1MUK
 
 | | reovirus lambda3 native structure | | Descriptor: | MINOR CORE PROTEIN LAMBDA 3 | | Authors: | Tao, Y, Farsetta, D.L, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2002-09-24 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA Synthesis in a Cage-Structural Studies of Reovirus Polymerase lambda3
Cell(Cambridge,Mass.), 111, 2002
|
|
1AW7
 
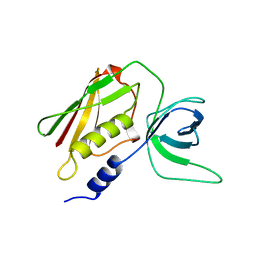 | | Q136A MUTANT OF TOXIC SHOCK SYNDROME TOXIN-1 FROM S. AUREUS | | Descriptor: | TOXIC SHOCK SYNDROME TOXIN-1 | | Authors: | Earhart, C.A, Mitchell, D.T, Murray, D.L, Pinheiro, D.M, Matsumura, M, Schlievert, P.M, Ohlendorf, D.H. | | Deposit date: | 1997-10-11 | | Release date: | 1998-10-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of five mutants of toxic shock syndrome toxin-1 with reduced biological activity.
Biochemistry, 37, 1998
|
|
1MWH
 
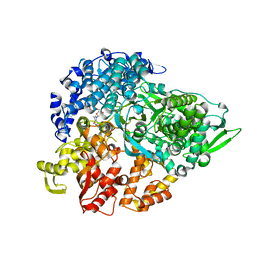 | | REOVIRUS POLYMERASE LAMBDA3 BOUND TO MRNA CAP ANALOG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, MANGANESE (II) ION, MINOR CORE PROTEIN LAMBDA 3 | | Authors: | Tao, Y, Farsetta, D.L, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2002-09-29 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA Synthesis in a Cage-Structural Studies of Reovirus Polymerase lambda3
Cell(Cambridge,Mass.), 111, 2002
|
|
1N38
 
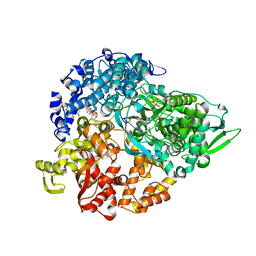 | | reovirus polymerase lambda3 elongation complex with one phosphodiester bond formed | | Descriptor: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, 3'-DEOXY-URIDINE 5'-TRIPHOSPHATE, 5'-R(*AP*UP*UP*AP*GP*C)-3', ... | | Authors: | Tao, Y, Farsetta, D.L, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2002-10-25 | | Release date: | 2002-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | RNA Synthesis in a Cage--Structural Studies of Reovirus Polymerase [lambda] 3
Cell(Cambridge,Mass.), 111, 2002
|
|
6OQO
 
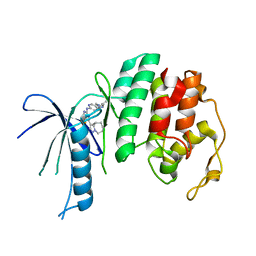 | | CDK6 in complex with Cpd24 N-(5-(6-ethyl-2,6-diazaspiro[3.3]heptan-2-yl)pyridin-2-yl)-5-fluoro-4-(4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl)pyrimidin-2-amine | | Descriptor: | Cyclin-dependent kinase 6, N-[5-(6-ethyl-2,6-diazaspiro[3.3]heptan-2-yl)pyridin-2-yl]-5-fluoro-4-[(4R)-4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl]pyrimidin-2-amine | | Authors: | Murray, J.M, Boenig, G.D.L. | | Deposit date: | 2019-04-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Design of a brain-penetrant CDK4/6 inhibitor for glioblastoma.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
1N35
 
 | | lambda3 elongation complex with four phosphodiester bond formed | | Descriptor: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, 5'-R(*AP*UP*UP*AP*GP*CP*CP*CP*CP*C)-3', 5'-R(P*GP*GP*GP*GP*G)-3', ... | | Authors: | Tao, Y, Farsetta, D.L, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2002-10-25 | | Release date: | 2002-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA Synthesis in a Cage--Structural Studies of Reovirus Polymerase [lambda] 3
Cell(Cambridge,Mass.), 111, 2002
|
|
6PBO
 
 | | Staphylococcus aureus Dihydrofolate reductase in complex with NADPH and UCP1232 | | Descriptor: | (4-{6-[(2S)-4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl]-2H-1,3-benzodioxol-4-yl}phenyl)acetic acid, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Reeve, S.M, Wright, D.L. | | Deposit date: | 2019-06-14 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Toward Broad Spectrum Dihydrofolate Reductase Inhibitors Targeting Trimethoprim Resistant Enzymes Identified in Clinical Isolates of Methicillin ResistantStaphylococcus aureus.
Acs Infect Dis., 5, 2019
|
|
6PD2
 
 | |
1C2T
 
 | | NEW INSIGHTS INTO INHIBITOR DESIGN FROM THE CRYSTAL STRUCTURE AND NMR STUDIES OF E. COLI GAR TRANSFORMYLASE IN COMPLEX WITH BETA-GAR AND 10-FORMYL-5,8,10-TRIDEAZAFOLIC ACID. | | Descriptor: | 10-FORMYL-5,8,10-TRIDEAZAFOLIC ACID, GLYCINAMIDE RIBONUCLEOTIDE, GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Authors: | Greasley, S.E, Yamashita, M.M, Cai, H, Benkovic, S.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 1999-07-26 | | Release date: | 2000-01-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New insights into inhibitor design from the crystal structure and NMR studies of Escherichia coli GAR transformylase in complex with beta-GAR and 10-formyl-5,8,10-trideazafolic acid.
Biochemistry, 38, 1999
|
|
6PD1
 
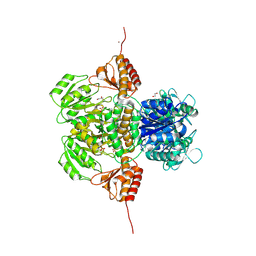 | |
6PF6
 
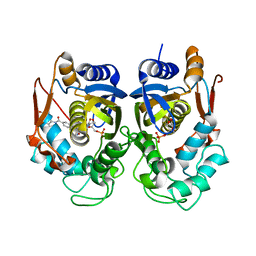 | | Crystal structure of TS-DHFR from Cryptosporidium hominis in complex with NADPH, FdUMP and 2-(4-((2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl)benzamido)terephthalic acid | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-({4-[(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzene-1-carbonyl}amino)benzene-1,4-dicarboxylic acid, Thymidylate synthase,Thymidylate synthase | | Authors: | Czyzyk, D.L, Valhondo, M, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2019-06-21 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structure activity relationship towards design of cryptosporidium specific thymidylate synthase inhibitors.
Eur.J.Med.Chem., 183, 2019
|
|
6P9Z
 
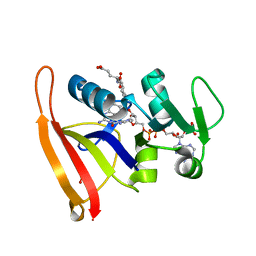 | |
1KJU
 
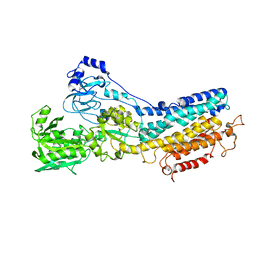 | | Ca2+-ATPase in the E2 State | | Descriptor: | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1a | | Authors: | Xu, C, Rice, W.J, He, W, Stokes, D.L. | | Deposit date: | 2001-12-05 | | Release date: | 2001-12-19 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | A structural model for the catalytic cycle of Ca(2+)-ATPase.
J.Mol.Biol., 316, 2002
|
|
8K55
 
 | |
8K57
 
 | |
1C3E
 
 | | NEW INSIGHTS INTO INHIBITOR DESIGN FROM THE CRYSTAL STRUCTURE AND NMR STUDIES OF E. COLI GAR TRANSFORMYLATE IN COMPLEX WITH BETA-GAR AND 10-FORMYL-5,8,10-TRIDEAZAFOLIC ACID. | | Descriptor: | 2-{4-[2-(2-AMINO-4-HYDROXY-QUINAZOLIN-6-YL)-1-CARBOXY-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, GLYCINAMIDE RIBONUCLEOTIDE, GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Authors: | Greasley, S.E, Yamashita, M.M, Cai, H, Benkovic, S.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 1999-07-27 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New insights into inhibitor design from the crystal structure and NMR studies of Escherichia coli GAR transformylase in complex with beta-GAR and 10-formyl-5,8,10-trideazafolic acid.
Biochemistry, 38, 1999
|
|
8OHN
 
 | | Human Coronavirus HKU1 spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Pronker, M.F, Hurdiss, D.L. | | Deposit date: | 2023-03-21 | | Release date: | 2023-08-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Sialoglycan binding triggers spike opening in a human coronavirus.
Nature, 624, 2023
|
|
8OPN
 
 | | Human Coronavirus HKU1 spike glycoprotein in complex with an alpha2,8-linked 9-O-acetylated disialoside (1-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 9-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid-(2-8)-N-acetyl-alpha-neuraminic acid, ... | | Authors: | Pronker, M.F, Creutznacher, R, Hurdiss, D.L. | | Deposit date: | 2023-04-07 | | Release date: | 2023-08-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Sialoglycan binding triggers spike opening in a human coronavirus.
Nature, 624, 2023
|
|
8OPO
 
 | | Human Coronavirus HKU1 spike glycoprotein in complex with an alpha2,8-linked 9-O-acetylated disialoside (3-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 9-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid-(2-8)-N-acetyl-alpha-neuraminic acid, ... | | Authors: | Pronker, M.F, Creutznacher, R, Hurdiss, D.L. | | Deposit date: | 2023-04-07 | | Release date: | 2023-08-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Sialoglycan binding triggers spike opening in a human coronavirus.
Nature, 624, 2023
|
|
8OPM
 
 | | Human Coronavirus HKU1 spike glycoprotein in complex with an alpha2,8-linked 9-O-acetylated disialoside (closed state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 9-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid-(2-8)-N-acetyl-alpha-neuraminic acid, ... | | Authors: | Pronker, M.F, Creutznacher, R, Hurdiss, D.L. | | Deposit date: | 2023-04-07 | | Release date: | 2023-08-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Sialoglycan binding triggers spike opening in a human coronavirus.
Nature, 624, 2023
|
|
1A2I
 
 | | SOLUTION STRUCTURE OF DESULFOVIBRIO VULGARIS (HILDENBOROUGH) FERROCYTOCHROME C3, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Messias, A.C, Kastrau, D.H.K, Costa, H.S, Legall, J, Turner, D.L, Santos, H, Xavier, A.V. | | Deposit date: | 1998-01-05 | | Release date: | 1998-07-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Desulfovibrio vulgaris (Hildenborough) ferrocytochrome c3: structural basis for functional cooperativity.
J.Mol.Biol., 281, 1998
|
|
1DSX
 
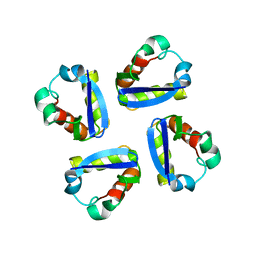 | | KV1.2 T1 DOMAIN, RESIDUES 33-119, T46V MUTANT | | Descriptor: | PROTEIN (KV1.2 VOLTAGE-GATED POTASSIUM CHANNEL) | | Authors: | Minor Jr, D.L, Lin, Y.-F, Mobley, B.C, Avelar, A, Jan, Y.N, Jan, L.Y, Berger, J.M. | | Deposit date: | 2000-01-10 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The polar T1 interface is linked to conformational changes that open the voltage-gated potassium channel.
Cell(Cambridge,Mass.), 102, 2000
|
|
1CIV
 
 | | CHLOROPLAST NADP-DEPENDENT MALATE DEHYDROGENASE FROM FLAVERIA BIDENTIS | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-MALATE DEHYDROGENASE | | Authors: | Carr, P.D, Ashton, A.R, Verger, D, Ollis, D.L. | | Deposit date: | 1999-04-01 | | Release date: | 2000-04-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Chloroplast NADP-malate dehydrogenase: structural basis of light-dependent regulation of activity by thiol oxidation and reduction.
Structure Fold.Des., 7, 1999
|
|
1EIA
 
 | |
5TBW
 
 | | Crystal structure of chlorolissoclimide bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Konst, Z.A, Szklarski, A.R, Pellegrino, S, Michalak, S.E, Meyer, M, Zanette, C, Cencic, R, Nam, S, Horne, D.A, Pelletier, J, Mobley, D.L, Yusupova, G, Yusupov, M, Vanderwal, C.D. | | Deposit date: | 2016-09-13 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis facilitates an understanding of the structural basis for translation inhibition by the lissoclimides.
Nat Chem, 9, 2017
|
|
