3QRI
 
 | | The crystal structure of human abl1 kinase domain in complex with DCC-2036 | | Descriptor: | 4-[4-({[3-tert-butyl-1-(quinolin-6-yl)-1H-pyrazol-5-yl]carbamoyl}amino)-3-fluorophenoxy]-N-methylpyridine-2-carboxamide, SODIUM ION, Tyrosine-protein kinase ABL1 | | Authors: | Chan, W.W, Wise, S.C, Kaufman, M.D, Ahn, Y.M, Ensinger, C.L, Haack, T, Hood, M.M, Jones, J, Lord, J.W, Lu, W.P, Miller, D, Patt, W.C, Smith, B.D, Petillo, P.A, Rutkoski, T.J, Telikepalli, H, Vogeti, L, Yao, T, Chun, L, Clark, R, Evangelista, P, Gavrilescu, L.C, Lazarides, K, Zaleskas, V.M, Stewart, L.J, Van Etten, R.A, Flynn, D.L. | | Deposit date: | 2011-02-18 | | Release date: | 2011-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational Control Inhibition of the BCR-ABL1 Tyrosine Kinase, Including the Gatekeeper T315I Mutant, by the Switch-Control Inhibitor DCC-2036.
Cancer Cell, 19, 2011
|
|
3QAZ
 
 | | IL-2 mutant D10 ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit gamma, Interleukin-2, ... | | Authors: | Levin, A.M, Bates, D.L, Ring, A.M, Lin, J.T, Su, L, Krieg, C, Bowman, G.R, Novick, P, Pande, V.S, Khort, H.E, Boyman, O, Gathman, C.G, Garcia, K.C. | | Deposit date: | 2011-01-12 | | Release date: | 2012-04-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.802 Å) | | Cite: | Exploiting a natural conformational switch to engineer an interleukin-2 'superkine'
Nature, 484, 2012
|
|
3QRJ
 
 | | The crystal structure of human abl1 kinase domain T315I mutant in complex with DCC-2036 | | Descriptor: | 4-[4-({[3-tert-butyl-1-(quinolin-6-yl)-1H-pyrazol-5-yl]carbamoyl}amino)-3-fluorophenoxy]-N-methylpyridine-2-carboxamide, Tyrosine-protein kinase ABL1 | | Authors: | Chan, W.W, Wise, S.C, Kaufman, M.D, Ahn, Y.M, Ensinger, C.L, Haack, T, Hood, M.M, Jones, J, Lord, J.W, Lu, W.P, Miller, D, Patt, W.C, Smith, B.D, Petillo, P.A, Rutkoski, T.J, Telikepalli, H, Vogeti, L, Yao, T, Chun, L, Clark, R, Evangelista, P, Gavrilescu, L.C, Lazarides, K, Zaleskas, V.M, Stewart, L.J, Van Etten, R.A, Flynn, D.L. | | Deposit date: | 2011-02-18 | | Release date: | 2011-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Conformational Control Inhibition of the BCR-ABL1 Tyrosine Kinase, Including the Gatekeeper T315I Mutant, by the Switch-Control Inhibitor DCC-2036.
Cancer Cell, 19, 2011
|
|
2I2R
 
 | | Crystal structure of the KChIP1/Kv4.3 T1 complex | | Descriptor: | CALCIUM ION, Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 3, ... | | Authors: | Findeisen, F, Pioletti, M, Minor Jr, D.L. | | Deposit date: | 2006-08-16 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Three-dimensional structure of the KChIP1-Kv4.3 T1 complex reveals a cross-shaped octamer
Nat.Struct.Mol.Biol., 13, 2006
|
|
2I5N
 
 | | 1.96 A X-ray structure of photosynthetic reaction center from Rhodopseudomonas viridis:Crystals grown by microfluidic technique | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Li, L, Mustafi, D, Fu, Q, Tereshko, V, Chen, D.L, Tice, J.D, Ismagilov, R.F. | | Deposit date: | 2006-08-25 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Nanoliter microfluidic hybrid method for simultaneous screening and optimization validated with crystallization of membrane proteins.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2I2H
 
 | | NMR structure of TPC3 in TFE | | Descriptor: | signaling peptide TCP3 | | Authors: | Syvitski, R.T, Jakeman, D.L, Li, Y. | | Deposit date: | 2006-08-16 | | Release date: | 2006-10-17 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure-Activity Analysis of Quorum-Sensing Signaling Peptides from Streptococcus mutans.
J.Bacteriol., 189, 2007
|
|
2EST
 
 | |
2H7Y
 
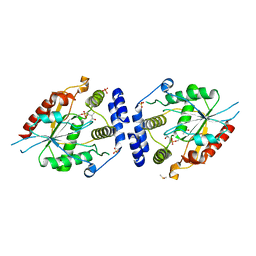 | | Pikromycin Thioesterase with covalent affinity label | | Descriptor: | DIMETHYL SULFOXIDE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Giraldes, J.W, Akey, D.L, Kittendorf, J.D, Sherman, D.H, Smith, J.S, Fecik, R.A. | | Deposit date: | 2006-06-06 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mechanistic Insights of Polyketide Macrolactonization from Polyketide-based Affinity Labels
NAT.CHEM.BIOL., 2, 2006
|
|
2HFJ
 
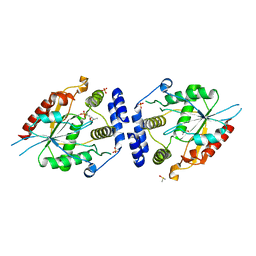 | | Pikromycin thioesterase with covalent pentaketide affinity label | | Descriptor: | DIMETHYL SULFOXIDE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Akey, D.L, Kittendorf, J.D, Giraldes, J.W, Fecik, R.A, Sherman, D.H, Smith, J.L. | | Deposit date: | 2006-06-24 | | Release date: | 2006-09-19 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for Macrolactonization by the Pikromycin Thioesterase
NAT.CHEM.BIOL., 2, 2006
|
|
2IYN
 
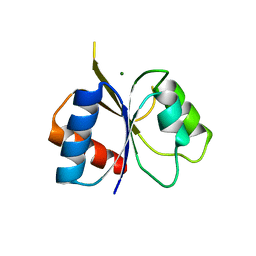 | | The co-factor-induced pre-active conformation in PhoB | | Descriptor: | MAGNESIUM ION, PHOSPHATE REGULON TRANSCRIPTIONAL REGULATORY PROTEIN PHOB | | Authors: | Sola, M, Drew, D.L, Blanco, A.G, Gomis-Ruth, F.X, Coll, M. | | Deposit date: | 2006-07-19 | | Release date: | 2006-08-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The Cofactor-Induced Pre-Active Conformation in Phob.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2J3H
 
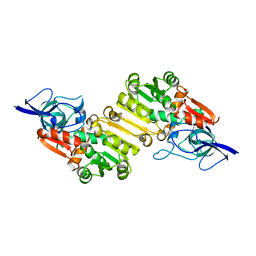 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Apo form | | Descriptor: | NADP-DEPENDENT OXIDOREDUCTASE P1 | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic and Structural Studies of Apoform, Binary, and Ternary Complexes of the Arabidopsis Alkenal Double Bond Reductase at5G16970.
J.Biol.Chem., 281, 2006
|
|
2J3K
 
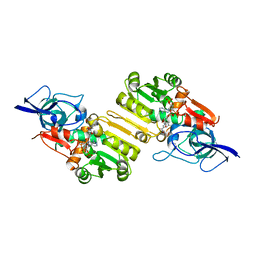 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Ternary Complex II | | Descriptor: | (2E,4R)-4-HYDROXYNON-2-ENAL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-dependent oxidoreductase 2-alkenal reductase | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-22 | | Release date: | 2006-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and structural studies of apoform, binary, and ternary complexes of the Arabidopsis alkenal double bond reductase At5g16970.
J. Biol. Chem., 281, 2006
|
|
2J3J
 
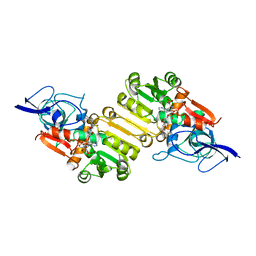 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Ternary Complex I | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-dependent oxidoreductase 2-alkenal reductase | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and structural studies of apoform, binary, and ternary complexes of the Arabidopsis alkenal double bond reductase At5g16970.
J. Biol. Chem., 281, 2006
|
|
2J3I
 
 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Binary Complex | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-DEPENDENT OXIDOREDUCTASE P1 | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and Structural Studies of Apoform, Binary, and Ternary Complexes of the Arabidopsis Alkenal Double Bond Reductase at5G16970.
J.Biol.Chem., 281, 2006
|
|
2FSU
 
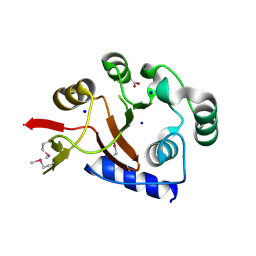 | | Crystal Structure of the PhnH Protein from Escherichia Coli | | Descriptor: | ACETATE ION, Protein phnH, SODIUM ION | | Authors: | Adams, M.A, Luo, Y, Zechel, D.L, Jia, Z, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-23 | | Release date: | 2007-03-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of PhnH: an essential component of carbon-phosphorus lyase in Escherichia coli.
J.Bacteriol., 190, 2008
|
|
2IU3
 
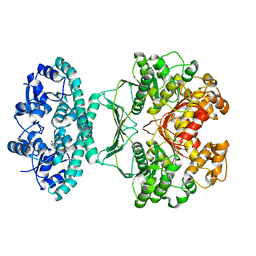 | | Crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 1,5-DIHYDROIMIDAZO[4,5-C][1,2,6]THIADIAZIN-4(3H)-ONE 2,2-DIOXIDE, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, POTASSIUM ION | | Authors: | Xu, L, Chong, Y, Hwang, I, D'Onofrio, A, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2006-05-27 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based design, synthesis, evaluation, and crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase.
J. Biol. Chem., 282, 2007
|
|
2IU0
 
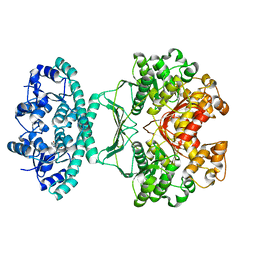 | | crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 1,5-DIHYDROIMIDAZO[4,5-C][1,2,6]THIADIAZIN-4(3H)-ONE 2,2-DIOXIDE, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, POTASSIUM ION | | Authors: | Xu, L, Chong, Y, Hwang, I, Onofrio, A.D, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2006-05-26 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure-Based Design, Synthesis, Evaluation, and Crystal Structures of Transition State Analogue Inhibitors of Inosine Monophosphate Cyclohydrolase
J.Biol.Chem., 282, 2007
|
|
2FRC
 
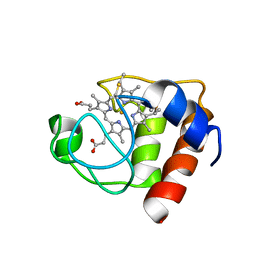 | | CYTOCHROME C (REDUCED) FROM EQUUS CABALLUS, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Qi, P.X, Di Stefano, D.L, Wand, A.J. | | Deposit date: | 1996-07-02 | | Release date: | 1997-07-29 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of horse heart ferricytochrome c and detection of redox-related structural changes by high-resolution 1H NMR.
Biochemistry, 35, 1996
|
|
2JHE
 
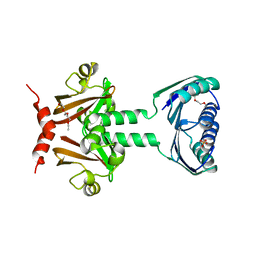 | | N-terminal domain of TyrR transcription factor (residues 1 - 190) | | Descriptor: | 2-(2-ETHOXYETHOXY)ETHANOL, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Verger, D, Carr, P.D, Kwok, T, Ollis, D.L. | | Deposit date: | 2007-02-21 | | Release date: | 2008-06-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the N-Terminal Domain of the Tyrr Transcription Factor Responsible for Gene Regulation of Aromatic Amino Acid Biosynthesis and Transport in Escherichia Coli K12
J.Mol.Biol., 367, 2007
|
|
2I2J
 
 | | NMR structure of UA159sp in TFE | | Descriptor: | Competence stimulating peptide | | Authors: | Syvitski, R.T, Jakeman, D.L, Li, Y. | | Deposit date: | 2006-08-16 | | Release date: | 2006-10-17 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure-Activity Analysis of Quorum-Sensing Signaling Peptides from Streptococcus mutans.
J.Bacteriol., 189, 2007
|
|
2K3V
 
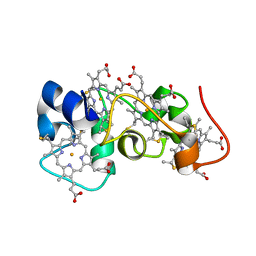 | | Solution Structure of a Tetrahaem Cytochrome from Shewanella Frigidimarina | | Descriptor: | HEME C, Tetraheme cytochrome c-type | | Authors: | Paixao, V.B, Turner, D.L, Salgueiro, C.A, Brennan, L, Reid, G.A, Chapman, S.K. | | Deposit date: | 2008-05-19 | | Release date: | 2009-03-31 | | Last modified: | 2019-10-02 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a tetraheme cytochrome from Shewanella frigidimarina reveals a novel family structural motif
Biochemistry, 47, 2008
|
|
2HFK
 
 | | Pikromycin thioesterase in complex with product 10-deoxymethynolide | | Descriptor: | (3R,4S,5S,7R,9E,11R,12R)-12-ETHYL-4-HYDROXY-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Akey, D.L, Kittendorf, J.D, Giraldes, J.W, Fecik, R.A, Sherman, D.H, Smith, J.L. | | Deposit date: | 2006-06-24 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis for Macrolactonization by the Pikromycin Thioesterase
NAT.CHEM.BIOL., 2, 2006
|
|
2GNK
 
 | | GLNK, A SIGNAL PROTEIN FROM E. COLI | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PROTEIN (NITROGEN REGULATORY PROTEIN) | | Authors: | Xu, Y, Cheah, E, Carr, P.D, van Heeswijk, W.C, Westerhoff, H.V, Vasudevan, S.G, Ollis, D.L. | | Deposit date: | 1998-07-14 | | Release date: | 1999-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | GlnK, a PII-homologue: structure reveals ATP binding site and indicates how the T-loops may be involved in molecular recognition.
J.Mol.Biol., 282, 1998
|
|
2GUI
 
 | | Structure and Function of Cyclized Versions of the Proofreading Exonuclease Subunit of E. coli DNA Polymerase III | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA polymerase III epsilon subunit, ... | | Authors: | Park, A.Y, Carr, P.D, Ollis, D.L, Dixon, N.E. | | Deposit date: | 2006-04-30 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Function of Cyclized Versions of the Proofreading Exonuclease Subunit E. coli DNA Polymerase III
To be Published
|
|
2GYS
 
 | | 2.7 A structure of the extracellular domains of the human beta common receptor involved in IL-3, IL-5, and GM-CSF signalling | | Descriptor: | Cytokine receptor common beta chain, alpha-L-fucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Carr, P.D, Conlan, F, Ford, S, Ollis, D.L, Young, I.G. | | Deposit date: | 2006-05-09 | | Release date: | 2006-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An improved resolution structure of the human beta common receptor involved in IL-3, IL-5 and GM-CSF signalling which gives better definition of the high-affinity binding epitope.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
