7SLR
 
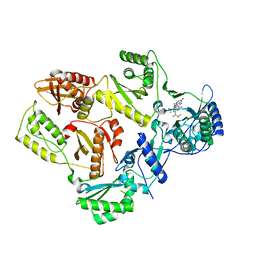 | | HIV Reverse Transcriptase with compound Pyr01 | | Descriptor: | 5-(difluoromethyl)-3-({1-[(5-fluoro-2-oxo-1,2-dihydropyridin-3-yl)methyl]-6-oxo-4-(1,1,2,2-tetrafluoroethyl)-1,6-dihydropyrimidin-5-yl}oxy)-2-methylbenzonitrile, Reverse transcriptase/ribonuclease H | | Authors: | Klein, D.J, Zebisch, M, Gu, M. | | Deposit date: | 2021-10-24 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.179 Å) | | Cite: | Potent targeted activator of cell kill molecules eliminate cells expressing HIV-1.
Sci Transl Med, 15, 2023
|
|
7FBP
 
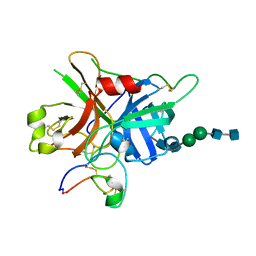 | | FXIIa-cMCoFx1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XIIa light chain, cMCoFx1 | | Authors: | Sengoku, T, Liu, W, de Veer, S.J, Huang, Y.H, Okada, C, Zdenek, C.N, Fry, B.G, Swedberg, J.E, Passioura, T, Craik, D.J, Suga, H, Ogata, K. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | An Ultrapotent and Selective Cyclic Peptide Inhibitor of Human beta-Factor XIIa in a Cyclotide Scaffold.
J.Am.Chem.Soc., 143, 2021
|
|
7JGX
 
 | |
7JHF
 
 | |
7JGY
 
 | |
6IEJ
 
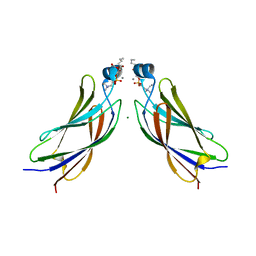 | | The C2 domain of cytosolic phospholipase A2 alpha bound to phosphatidylcholine | | Descriptor: | 1,2-dihexanoyl-sn-glycero-3-phosphocholine, CALCIUM ION, Cytosolic phospholipase A2, ... | | Authors: | Hirano, Y, Gao, Y.G, Stephenson, D.J, Vu, N.T, Malinina, L, Chalfant, C.E, Patel, D.J, Brown, R.E. | | Deposit date: | 2018-09-14 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structural basis of phosphatidylcholine recognition by the C2-domain of cytosolic phospholipase A2alpha.
Elife, 8, 2019
|
|
1W8C
 
 | |
8BMQ
 
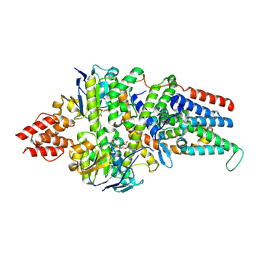 | | Cryo-EM structure of the folate-specific ECF transporter complex in MSP2N2 lipid nanodiscs bound to AMP-PNP | | Descriptor: | Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, Energy-coupling factor transporter transmembrane protein EcfT, ... | | Authors: | Thangaratnarajah, C, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2022-11-10 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Expulsion mechanism of the substrate-translocating subunit in ECF transporters.
Nat Commun, 14, 2023
|
|
8BMS
 
 | | Cryo-EM structure of the mutant solitary ECF module 2EQ in MSP2N2 lipid nanodiscs in the ATPase closed and ATP-bound conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, ... | | Authors: | Thangaratnarajah, C, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2022-11-10 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Expulsion mechanism of the substrate-translocating subunit in ECF transporters.
Nat Commun, 14, 2023
|
|
8BMP
 
 | | Cryo-EM structure of the folate-specific ECF transporter complex in MSP2N2 lipid nanodiscs bound to ATP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Energy-coupling factor transporter ATP-binding protein EcfA1, ... | | Authors: | Thangaratnarajah, C, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2022-11-10 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Expulsion mechanism of the substrate-translocating subunit in ECF transporters.
Nat Commun, 14, 2023
|
|
8BMR
 
 | | Cryo-EM structure of the wild-type solitary ECF module in MSP2N2 lipid nanodiscs in the ATPase open and nucleotide-free conformation | | Descriptor: | Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, Energy-coupling factor transporter transmembrane protein EcfT | | Authors: | Thangaratnarajah, C, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2022-11-10 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Expulsion mechanism of the substrate-translocating subunit in ECF transporters.
Nat Commun, 14, 2023
|
|
8I6N
 
 | | Crystal structure of Co-type nitrile hydratase mutant from Pseudomonas thermophila - L6T | | Descriptor: | COBALT (II) ION, Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta | | Authors: | Ma, D, Cheng, Z.Y, Lai, Q.P, Hou, X.D, Yin, D.J, Rao, Y.J, Zhou, Z.M. | | Deposit date: | 2023-01-29 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Co-type nitrile hydratase mutant L6T from Pseudomonas thermophila at 2.2 Angstroms resolution.
To Be Published
|
|
5VMP
 
 | | Crystal Structure of Human KDM4 with Small Molecule Inhibitor QC5714 | | Descriptor: | 3-({[(1R)-6-methoxy-1,2,3,4-tetrahydronaphthalen-1-yl]methyl}amino)pyridine-4-carboxylic acid, Lysine-specific demethylase 4A, NICKEL (II) ION, ... | | Authors: | Hosfield, D.J. | | Deposit date: | 2017-04-28 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Design of KDM4 Inhibitors with Antiproliferative Effects in Cancer Models.
ACS Med Chem Lett, 8, 2017
|
|
5VN1
 
 | | horse liver alcohol dehydrogenae complexed with NADH (R,S)-N-1-methylhexylformamide | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (R)-N-(1-METHYL-HEXYL)-FORMAMIDE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Plapp, B.V, Ramaswamy, S, Ferraro, D.J, Baskar Raj, S. | | Deposit date: | 2017-04-28 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Horse Liver Alcohol Dehydrogenase: Zinc Coordination and Catalysis.
Biochemistry, 56, 2017
|
|
5VW1
 
 | | Crystal structure of SpyCas9-sgRNA-AcrIIA4 ternary complex | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9/Csn1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, H, Patel, D.J. | | Deposit date: | 2017-05-21 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Inhibition Mechanism of an Anti-CRISPR Suppressor AcrIIA4 Targeting SpyCas9.
Mol. Cell, 67, 2017
|
|
7T69
 
 | | Crystal structure of Avr3 (SIX1) from Fusarium oxysporum f. sp. lycopersici | | Descriptor: | Avr3 (SIX1), Secreted in xylem 1, SULFATE ION | | Authors: | Yu, D.S, Outram, M.A, Ericsson, D.J, Jones, D.A, Williams, S.J. | | Deposit date: | 2021-12-13 | | Release date: | 2023-01-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The structural repertoire of Fusarium oxysporum f. sp. lycopersici effectors revealed by experimental and computational studies
Elife, 2023
|
|
7T6A
 
 | | Crystal structure of Avr1 (SIX4) from Fusarium oxysporum f. sp. lycopersici | | Descriptor: | Avr1 (FolSIX4), Avirulence protein 1, Avr1 (SIX4), ... | | Authors: | Yu, D.S, Outram, M.A, Ericsson, D.J, Jones, D.A, Williams, S.J. | | Deposit date: | 2021-12-13 | | Release date: | 2023-01-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structural repertoire of Fusarium oxysporum f. sp. lycopersici effectors revealed by experimental and computational studies
Elife, 2023
|
|
1HR2
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A MUTANT P4-P6 DOMAIN (DELC209) OF TETRAHYMENA THEMOPHILA GROUP I INTRON. | | Descriptor: | MAGNESIUM ION, P4-P6 DELC209 MUTANT RNA RIBOZYME DOMAIN | | Authors: | Juneau, K, Podell, E.R, Harrington, D.J, Cech, T.R. | | Deposit date: | 2000-12-20 | | Release date: | 2001-04-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA--solvent interactions.
Structure, 9, 2001
|
|
1HYK
 
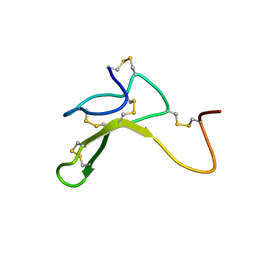 | | AGOUTI-RELATED PROTEIN (87-132) (AC-AGRP(87-132)) | | Descriptor: | AGOUTI RELATED PROTEIN | | Authors: | Bolin, K.A, Anderson, D.J, Trulson, J.A, Thompson, D.A, Wilken, J, Kent, S.B.H, Millhauser, G.L. | | Deposit date: | 2001-01-19 | | Release date: | 2001-02-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a minimized human agouti related protein prepared by total chemical synthesis.
FEBS Lett., 451, 1999
|
|
1HVZ
 
 | |
1I2Z
 
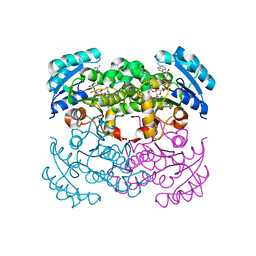 | | E. COLI ENOYL REDUCTASE IN COMPLEX WITH NAD AND BRL-12654 | | Descriptor: | 4-(2-THIENYL)-1-(4-METHYLBENZYL)-1H-IMIDAZOLE, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Heerding, D.A, Miller, W.H, Payne, D.J, Janson, C.A, Qiu, X. | | Deposit date: | 2001-02-12 | | Release date: | 2002-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 1,4-Disubstituted imidazoles are potential antibacterial agents functioning as inhibitors of enoyl acyl carrier protein
reductase (FabI).
Bioorg.Med.Chem.Lett., 11, 2001
|
|
1HY7
 
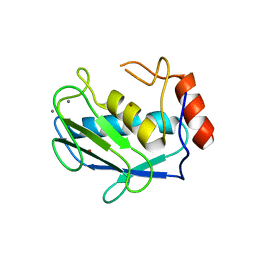 | | A CARBOXYLIC ACID BASED INHIBITOR IN COMPLEX WITH MMP3 | | Descriptor: | CALCIUM ION, R-2-{[4'-METHOXY-(1,1'-BIPHENYL)-4-YL]-SULFONYL}-AMINO-6-METHOXY-HEX-4-YNOIC ACID, STROMELYSIN-1, ... | | Authors: | Natchus, M.G, Bookland, R.G, Laufersweiler, M.J, Pikul, S, Almstead, N.G, De, B, Janusz, M.J, Hsieh, L.C, Gu, F, Pokross, M.E, Patel, V.S, Garver, S.M, Peng, S.X, Branch, T.M, King, S.L, Baker, T.R, Foltz, D.J, Mieling, G.E. | | Deposit date: | 2001-01-18 | | Release date: | 2002-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Development of new carboxylic acid-based MMP inhibitors derived from functionalized propargylglycines.
J.Med.Chem., 44, 2001
|
|
1I3A
 
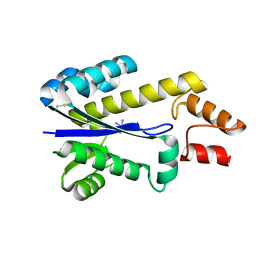 | | RNASE HII FROM ARCHAEOGLOBUS FULGIDUS WITH COBALT HEXAMMINE CHLORIDE | | Descriptor: | COBALT HEXAMMINE(III), RIBONUCLEASE HII | | Authors: | Chapados, B.R, Chai, Q, Hosfield, D.J, Qiu, J, Shen, B, Tainer, J.A. | | Deposit date: | 2001-02-13 | | Release date: | 2001-04-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural biochemistry of a type 2 RNase H: RNA primer recognition and removal during DNA replication.
J.Mol.Biol., 307, 2001
|
|
1I30
 
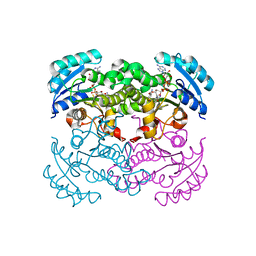 | | E. Coli Enoyl Reductase +NAD+SB385826 | | Descriptor: | 1,3,4,9-TETRAHYDRO-2-(HYDROXYBENZOYL)-9-[(4-HYDROXYPHENYL)METHYL]-6-METHOXY-2H-PYRIDO[3,4-B]INDOLE, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Seefeld, M.A, Miller, W.H, Payne, D.J, Janson, C.A, Qiu, X. | | Deposit date: | 2001-02-12 | | Release date: | 2002-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibitors of bacterial enoyl acyl carrier protein reductase (FabI): 2,9-disubstituted 1,2,3,4-tetrahydropyrido[3,4-b]indoles as potential antibacterial agents.
Bioorg.Med.Chem.Lett., 11, 2001
|
|
1I34
 
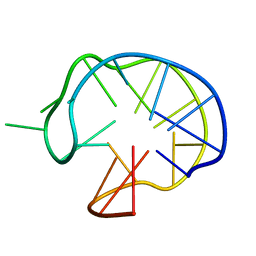 | | SOLUTION DNA QUADRUPLEX WITH DOUBLE CHAIN REVERSAL LOOP AND TWO DIAGONAL LOOPS CONNECTING GGGG TETRADS FLANKED BY G-(T-T) TRIAD AND T-T-T TRIPLE | | Descriptor: | 5'-D(*GP*GP*TP*TP*TP*TP*GP*GP*CP*AP*GP*GP*GP*TP*TP*TP*TP*GP*GP*T)-3' | | Authors: | Kuryavyi, V, Majumdar, A, Shallop, A, Chernichenko, N, Skripkin, E, Jones, R, Patel, D.J. | | Deposit date: | 2001-02-12 | | Release date: | 2001-06-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A double chain reversal loop and two diagonal loops define the architecture of a unimolecular DNA quadruplex containing a pair of stacked G(syn)-G(syn)-G(anti)-G(anti) tetrads flanked by a G-(T-T) Triad and a T-T-T triple.
J.Mol.Biol., 310, 2001
|
|
