8P0L
 
 | | Crystal structure of human O-GlcNAcase in complex with an S-linked CKII peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYSTEINE, Protein O-GlcNAcase, ... | | Authors: | Males, A, Davies, G.J, Calvelo, M, Alteen, M.G, Vocadlo, D.J, Rovira, C. | | Deposit date: | 2023-05-10 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human O -GlcNAcase Uses a Preactivated Boat-skew Substrate Conformation for Catalysis. Evidence from X-ray Crystallography and QM/MM Metadynamics.
Acs Catalysis, 13, 2023
|
|
8QQF
 
 | | TBC1D23 PH domain complexed with STX16 TLY motif | | Descriptor: | GLYCEROL, PHOSPHATE ION, Syntaxin-16, ... | | Authors: | Kaufman, J.G, Owen, D.J. | | Deposit date: | 2023-10-04 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Cargo selective vesicle tethering: The structural basis for binding of specific cargo proteins by the Golgi tether component TBC1D23.
Sci Adv, 10, 2024
|
|
8QZE
 
 | | Heme-domain BM3 variant 21B3_F87V-A328F | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, IMIDAZOLE, ... | | Authors: | Opperman, D.J, Ebrecht, A.C, Aschenbrenner, J.C. | | Deposit date: | 2023-10-27 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Revisiting strategies and their combinatorial effect for introducing peroxygenase activity in CYP102A1 (P450BM3)
Mol Catal, 557, 2024
|
|
8RGI
 
 | | Structure of DYNLT1:DYNLT2B (TCTEX1:TCTEX1D2) heterodimer. | | Descriptor: | Dynein light chain Tctex-type 1, Dynein light chain Tctex-type protein 2B | | Authors: | Mukhopadhyay, A.G, Toropova, K, Daly, L, Wells, J, Vuolo, L, Seda, M, Jenkins, D, Stephens, D.J, Roberts, A.J. | | Deposit date: | 2023-12-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure and tethering mechanism of dynein-2 intermediate chains in intraflagellar transport.
Embo J., 43, 2024
|
|
8QZF
 
 | | Heme-domain BM3 mutant T268E | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional cytochrome P450/NADPH--P450 reductase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Opperman, D.J, Ebrecht, A.C, Aschenbrenner, J.C. | | Deposit date: | 2023-10-27 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Revisiting strategies and their combinatorial effect for introducing peroxygenase activity in CYP102A1 (P450BM3)
Mol Catal, 557, 2024
|
|
8RGG
 
 | | Structure of dynein-2 intermediate chain DYNC2I2 (WDR34) in complex with dynein-2 heavy chain DYNC2H1. | | Descriptor: | Cytoplasmic dynein 2 intermediate chain 1, Cytoplasmic dynein 2 intermediate chain 2, Dynein light chain 1, ... | | Authors: | Mukhopadhyay, A.G, Toropova, K, Daly, L, Wells, J, Vuolo, L, Mladenov, M, Seda, M, Jenkins, D, Stephens, D.J, Roberts, A.J. | | Deposit date: | 2023-12-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure and tethering mechanism of dynein-2 intermediate chains in intraflagellar transport.
Embo J., 43, 2024
|
|
8RGH
 
 | | Structure of dynein-2 intermediate chain DYNC2I1 (WDR60) in complex with the dynein-2 heavy chain DYNC2H1. | | Descriptor: | Cytoplasmic dynein 2 intermediate chain 1, Cytoplasmic dynein 2 intermediate chain 2, Cytoplasmic dynein 2 light intermediate chain 1, ... | | Authors: | Mukhopadhyay, A.G, Toropova, K, Daly, L, Wells, J, Vuolo, L, Seda, M, Jenkins, D, Stephens, D.J, Roberts, A.J. | | Deposit date: | 2023-12-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure and tethering mechanism of dynein-2 intermediate chains in intraflagellar transport.
Embo J., 43, 2024
|
|
8Q5J
 
 | |
8QRQ
 
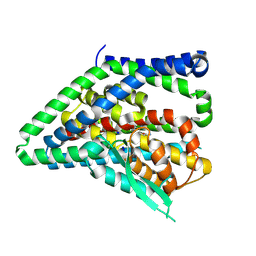 | | ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the outward-facing state (OFS.2) | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0), SODIUM ION | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
8QRW
 
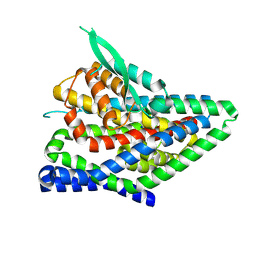 | | ASCT2 protomer in lipid nanodiscs under low Na+ concentration in the intermediate outward-facing state (iOFS-up) | | Descriptor: | Neutral amino acid transporter B(0), SODIUM ION | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
8QRR
 
 | | ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the outward-facing state (OFS.3) | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0), SODIUM ION | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
8QRP
 
 | | ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the outward-facing state (OFS.1) | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0), SODIUM ION | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
8QRU
 
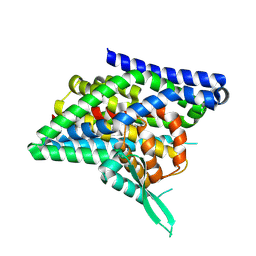 | | ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the intermediate outward-facing state (iOFS-down) | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0) | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
8QRS
 
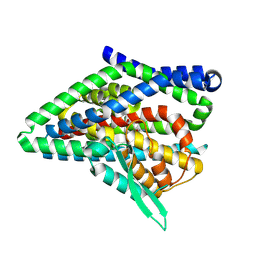 | | ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the intermediate outward-facing state (iOFS-up) | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0) | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
8QRO
 
 | | ASCT2 trimer in lipid nanodiscs with bound glutamine and Na+ ions in the outward-facing state (OFS) | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0), SODIUM ION | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
8QRV
 
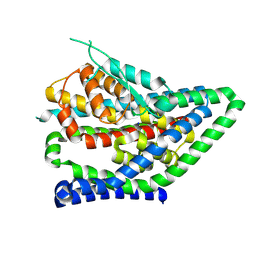 | | ASCT2 protomer in lipid nanodiscs under low Na+ concentration in the outward-facing state (OFS) | | Descriptor: | Neutral amino acid transporter B(0), SODIUM ION | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
4LTS
 
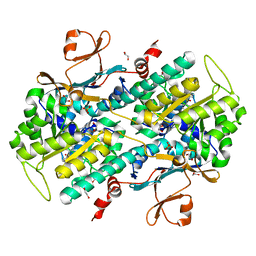 | | Discovery of Potent and Efficacious Cyanoguanidine-containing Nicotinamide Phosphoribosyltransferase (Nampt) Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 2-cyano-1-pyridin-4-yl-3-(4-{[3-(trifluoromethoxy)phenyl]sulfonyl}benzyl)guanidine, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Zheng, X, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Han, B, Ho, Y, Kley, N, Lin, J, Reynoids, D.J, Sharma, G, Smith, C.C, Wang, Z, Dragovich, P.S, Oh, A, Wang, W, Zak, M, Wang, L, Yuen, P, Bair, K.W. | | Deposit date: | 2013-07-23 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | Discovery of potent and efficacious cyanoguanidine-containing nicotinamide phosphoribosyltransferase (Nampt) inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3PTA
 
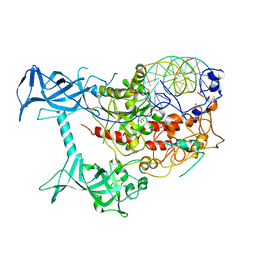 | | Crystal structure of human DNMT1(646-1600) in complex with DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*CP*GP*GP*AP*GP*GP*CP*TP*CP*AP*CP*GP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*CP*GP*TP*GP*AP*GP*CP*CP*TP*CP*CP*GP*CP*AP*GP*G)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation.
Science, 331, 2011
|
|
4G6B
 
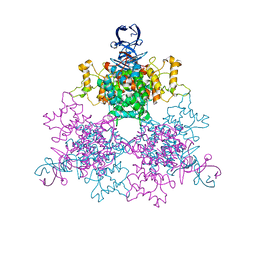 | | Three dimensional structure analysis of the type II citrate synthase from e.coli | | Descriptor: | Citrate synthase, SULFATE ION | | Authors: | Nguyen, N.T, Maurus, R, Stokell, D.J, Ayed, A, Duckworth, H.W, Brayer, G.D. | | Deposit date: | 2012-07-18 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparative Analysis of Folding and Substrate Binding Sites between Regulated Hexameric Type II Citrate Synthases and Unregulated Dimeric Type I Enzymes.
Biochemistry, 40, 2001
|
|
4GBA
 
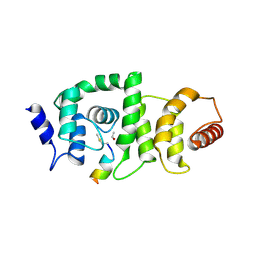 | | DCNL complex with N-terminally acetylated NEDD8 E2 peptide | | Descriptor: | DCN1-like protein 3, NEDD8-conjugating enzyme UBE2F | | Authors: | Monda, J.K, Scott, D.C, Miller, D.J, Harper, J.W, Bennett, E.J, Schulman, B.A. | | Deposit date: | 2012-07-26 | | Release date: | 2012-11-28 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Conservation of Distinctive N-terminal Acetylation-Dependent Interactions across a Family of Mammalian NEDD8 Ligation Enzymes.
Structure, 21, 2013
|
|
4FT4
 
 | |
8H5D
 
 | |
4X9X
 
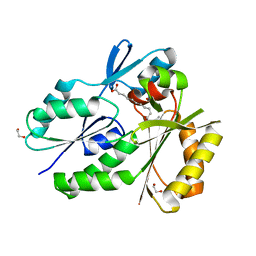 | | Biochemical Roles for Conserved Residues in the Bacterial Fatty Acid Binding Protein Family | | Descriptor: | 1,2-ETHANEDIOL, DegV domain-containing protein MW1315, OLEIC ACID | | Authors: | Broussard, T.C, Miller, D.J, Jackson, P, Nourse, A, Rock, C.O. | | Deposit date: | 2014-12-11 | | Release date: | 2016-01-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Biochemical Roles for Conserved Residues in the Bacterial Fatty Acid-binding Protein Family.
J.Biol.Chem., 291, 2016
|
|
4GAO
 
 | | DCNL complex with N-terminally acetylated NEDD8 E2 peptide | | Descriptor: | BROMIDE ION, DCN1-like protein 2, NEDD8-conjugating enzyme Ubc12 | | Authors: | Monda, J.K, Scott, D.C, Miller, D.J, Harper, J.W, Bennett, E.J, Schulman, B.A. | | Deposit date: | 2012-07-25 | | Release date: | 2012-11-28 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structural Conservation of Distinctive N-terminal Acetylation-Dependent Interactions across a Family of Mammalian NEDD8 Ligation Enzymes.
Structure, 21, 2013
|
|
4XD2
 
 | | Horse liver alcohol dehydrogenase-NADH complex | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Alcohol dehydrogenase E chain, ... | | Authors: | Plapp, B.V, Baskar Raj, S, Ferraro, D.J. | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of Horse liver alcohol dehydrogenase complexed with NADH
To Be Published
|
|
