8OVV
 
 | |
7MFT
 
 | | Glutamate synthase, glutamate dehydrogenase counter-enzyme complex (GudB6-GltA6-GltB6) | | Descriptor: | FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Jayaraman, V, Lee, D.J, Elad, N, Fraser, J.S, Tawfik, D.S. | | Deposit date: | 2021-04-11 | | Release date: | 2022-01-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | A counter-enzyme complex regulates glutamate metabolism in Bacillus subtilis.
Nat.Chem.Biol., 18, 2022
|
|
7MFM
 
 | | Glutamate synthase, glutamate dehydrogenase counter-enzyme complex | | Descriptor: | FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Jayaraman, V, Lee, D.J, Elad, N, Fraser, J.S, Tawfik, D.S. | | Deposit date: | 2021-04-10 | | Release date: | 2022-01-05 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | A counter-enzyme complex regulates glutamate metabolism in Bacillus subtilis.
Nat.Chem.Biol., 18, 2022
|
|
7MKK
 
 | |
7NGH
 
 | | Structure of glutamate transporter homologue in complex with Sybody | | Descriptor: | ASPARTIC ACID, Proton/glutamate symporter, SDF family, ... | | Authors: | Arkhipova, V, Slotboom, D.J, Guskov, A. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Kinetic mechanism of Na + -coupled aspartate transport catalyzed by Glt Tk .
Commun Biol, 4, 2021
|
|
8FKF
 
 | |
8FKD
 
 | |
8FKG
 
 | |
8FKC
 
 | |
8FKE
 
 | |
8FHE
 
 | |
8FHG
 
 | |
8FHF
 
 | |
8UPV
 
 | | Structure of SARS-Cov2 3CLPro in complex with Compound 33 | | Descriptor: | 3C-like proteinase nsp5, methyl {(2S)-1-[(1S,3aR,6aS)-1-{[(2R,3S,6R)-6-fluoro-2-hydroxy-1-(methylamino)-1-oxoheptan-3-yl]carbamoyl}hexahydrocyclopenta[c]pyrrol-2(1H)-yl]-3,3-dimethyl-1-oxobutan-2-yl}carbamate | | Authors: | Krishnamurthy, H, Zhuang, N, Qiang, D, Wu, Y, Klein, D.J. | | Deposit date: | 2023-10-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
8UPS
 
 | | Structure of SARS-Cov2 3CLPro in complex with Compound 5 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, PHOSPHATE ION | | Authors: | Wu, Y, Qiang, D, Zhuang, N, Krishnamurthy, H, Klein, D.J. | | Deposit date: | 2023-10-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
8ORX
 
 | |
8OS2
 
 | | Structure of the human LYVE-1 (lymphatic vessel endothelial receptor-1) hyaluronan binding domain in an unliganded state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Lymphatic vessel endothelial hyaluronic acid receptor 1 | | Authors: | Ni, T, Bannerji, S, Jackson, D.J, Gilbert, R.J.C. | | Deposit date: | 2023-04-17 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A novel sliding interaction with hyaluronan underlies the mode of action of LYVE-1, the receptor for leucocyte entry and trafficking in the lymphatics
To be published
|
|
4AE2
 
 | | Crystal structure of Human fibrillar procollagen type III C- propeptide trimer | | Descriptor: | CALCIUM ION, COLLAGEN ALPHA-1(III) CHAIN, NITRATE ION | | Authors: | Bourhis, J.M, Mariano, N, Zhao, Y, Harlos, K, Jones, E.Y, Moali, C, Aghajari, N, Hulmes, D.J. | | Deposit date: | 2012-01-05 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Basis of Fibrillar Collagen Trimerization and Related Genetic Disorders.
Nat.Struct.Mol.Biol., 19, 2012
|
|
8UAU
 
 | | human ATE1 in complex with Arg-tRNA and a peptide substrate | | Descriptor: | Isoform ATE1-2 of Arginyl-tRNA--protein transferase 1, RNA (76-MER), ZINC ION, ... | | Authors: | Huang, W, Zhang, Y, Taylor, D.J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Oligomerization and a distinct tRNA-binding loop are important regulators of human arginyl-transferase function.
Nat Commun, 15, 2024
|
|
8OF8
 
 | | Cryo-EM structure of actomyosin-5a-S1 with the full-length lever (nucleotide free) | | Descriptor: | Actin, alpha skeletal muscle, Calmodulin-1, ... | | Authors: | Gravett, M.S.C, Klebl, D.P, Harlen, O.G, Read, D.J, Harris, S.A, Muench, S.P, Peckham, M. | | Deposit date: | 2023-03-14 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Unveiling the Structural Dynamics of Myosin 5a: Cryo-EM Insights into the Motor Protein's Lever Conformations and Walking Mechanics
To Be Published
|
|
7LTG
 
 | | STRUCTURE OF HUMAN HDAC2 IN COMPLEX WITH APICIDIN | | Descriptor: | (3S,6S,9S,15aR)-9-[(2S)-butan-2-yl]-3-(6,6-dihydroxyoctyl)-6-[(1-methoxy-1H-indol-3-yl)methyl]octahydro-2H-pyrido[1,2-a][1,4,7,10]tetraazacyclododecine-1,4,7,10(3H,12H)-tetrone, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Beshore, D.C. | | Deposit date: | 2021-02-19 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Redefining the Histone Deacetylase Inhibitor Pharmacophore: High Potency with No Zinc Cofactor Interaction.
Acs Med.Chem.Lett., 12, 2021
|
|
7LTK
 
 | |
7LTL
 
 | |
3J48
 
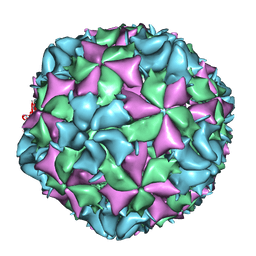 | | Cryo-EM structure of Poliovirus 135S particles | | Descriptor: | Protein VP1, Protein VP2, Protein VP3 | | Authors: | Butan, C, Fiman, D.J, Hogle, J.M. | | Deposit date: | 2013-06-28 | | Release date: | 2013-12-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Cryo-Electron Microscopy Reconstruction Shows Poliovirus 135S Particles Poised for Membrane Interaction and RNA Release.
J.Virol., 88, 2014
|
|
8FZR
 
 | |
