6U7W
 
 | | NMR solution structure of a triazole bridged KLK7 inhibitor | | Descriptor: | 1-methyl-1H-1,2,3-triazole, GLY-LYS-ALA-LEU-PHE-SER-ASN-PRO-PRO-ILE-ALA-PHE-PRO-ASN | | Authors: | White, A.M, Harvey, P.J, Durek, T, Craik, D.J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2020-07-15 | | Method: | SOLUTION NMR | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1M6P
 
 | | EXTRACYTOPLASMIC DOMAIN OF BOVINE CATION-DEPENDENT MANNOSE 6-PHOSPHATE RECEPTOR | | Descriptor: | 6-O-phosphono-alpha-D-mannopyranose, CATION-DEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR, MANGANESE (II) ION | | Authors: | Roberts, D.L, Weix, D.J, Dahms, N.M, Kim, J.J.-P. | | Deposit date: | 1998-04-19 | | Release date: | 1999-04-27 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis of lysosomal enzyme recognition: three-dimensional structure of the cation-dependent mannose 6-phosphate receptor.
Cell(Cambridge,Mass.), 93, 1998
|
|
4NJ5
 
 | | Crystal structure of SUVH9 | | Descriptor: | Probable histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH9, ZINC ION | | Authors: | Du, J, Patel, D.J. | | Deposit date: | 2013-11-08 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation.
Nature, 507, 2014
|
|
6UF1
 
 | | Pistol ribozyme transition-state analog vanadate | | Descriptor: | MANGANESE (II) ION, RNA (5'-R(*UP*CP*UP*GP*CP*UP*CP*UP*CP*(GVA)*UP*CP*CP*AP*A)-3'), RNA (50-MER) | | Authors: | Teplova, M, Falschlunger, C, Krasheninina, O, Patel, D.J, Micura, R. | | Deposit date: | 2019-09-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crucial Roles of Two Hydrated Mg2+Ions in Reaction Catalysis of the Pistol Ribozyme.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1M01
 
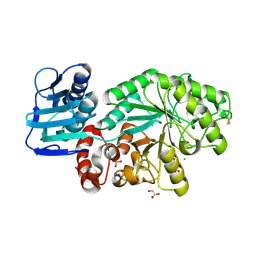 | | Wildtype Streptomyces plicatus beta-hexosaminidase in complex with product (GlcNAc) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | J Williams, S, Mark, B.L, Vocadlo, D.J, James, M.N.G, Withers, S.G. | | Deposit date: | 2002-06-11 | | Release date: | 2003-01-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Aspartate 313 in the Streptomyces plicatus hexosaminidase plays a critical
role in substrate-assisted catalysis by orienting the 2-acetamido group
and stabilizing the transition state.
J.Biol.Chem., 277, 2002
|
|
6O0R
 
 | | Crystal structure of the TIR domain from human SARM1 in complex with glycerol | | Descriptor: | GLYCEROL, Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Deerain, N, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
1MKO
 
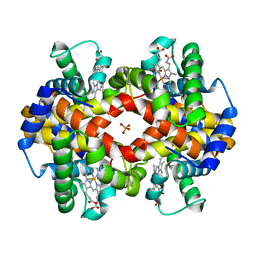 | |
6O0U
 
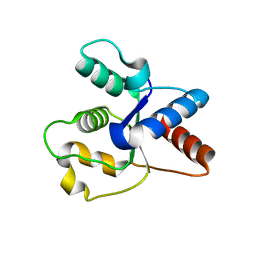 | | Crystal structure of the TIR domain H685A mutant from human SARM1 | | Descriptor: | Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6NXK
 
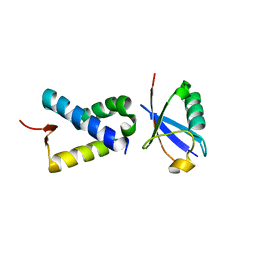 | | Ubiquitin binding variants | | Descriptor: | Anaphase-promoting complex subunit 2, Polyubiquitin-C | | Authors: | Miller, D.J, Watson, E.R. | | Deposit date: | 2019-02-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein engineering of a ubiquitin-variant inhibitor of APC/C identifies a cryptic K48 ubiquitin chain binding site.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3LPY
 
 | | Crystal structure of the RRM domain of CyP33 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Peptidyl-prolyl cis-trans isomerase E, SULFATE ION | | Authors: | Wang, Z, Patel, D.J. | | Deposit date: | 2010-02-07 | | Release date: | 2010-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pro isomerization in MLL1 PHD3-bromo cassette connects H3K4me readout to CyP33 and HDAC-mediated repression.
Cell(Cambridge,Mass.), 141, 2010
|
|
5AD1
 
 | | A complex of the synthetic siderophore analogue Fe(III)-8-LICAM with the CeuE periplasmic protein from Campylobacter jejuni | | Descriptor: | ENTEROCHELIN UPTAKE PERIPLASMIC BINDING PROTEIN, FE (III) ION, N,N'-OCTANE-1,8-DIYLBIS(2,3-DIHYDROXYBENZAMIDE) | | Authors: | Blagova, E, Hughes, A, Moroz, O.V, Raines, D.J, Wilde, E.J, Turkenburg, J.P, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2015-08-19 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
6O0Q
 
 | | Crystal structure of the TIR domain from human SARM1 in complex with ribose | | Descriptor: | CHLORIDE ION, Sterile alpha and TIR motif-containing protein 1, beta-D-ribofuranose | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Deerain, N, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
4APY
 
 | | Ethylene glycol-bound form of P450 CYP125A3 from Mycobacterium smegmatis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, P450 HEME-THIOLATE PROTEIN, ... | | Authors: | Frank, D.J, Garcia Fernandez, E, Kells, P.M, Garcia Lopez, J.L, Podust, L.M, Ortiz de Montellano, P.R. | | Deposit date: | 2012-04-09 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Highly Conserved Mycobacterial Cholesterol Catabolic Pathway.
Environ.Microbiol., 15, 2013
|
|
1M31
 
 | | Three-Dimensional Solution Structure of Apo-Mts1 | | Descriptor: | Placental calcium-binding protein | | Authors: | Vallely, K.M, Rustandi, R.R, Ellis, K.C, Varlamova, O, Bresnick, A.R, Weber, D.J. | | Deposit date: | 2002-06-26 | | Release date: | 2002-10-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human Mts1 (S100A4) as determined by NMR spectroscopy.
Biochemistry, 41, 2002
|
|
5ABH
 
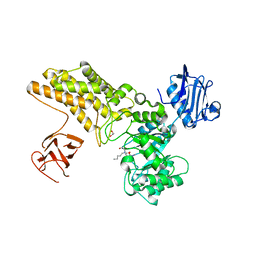 | | Structure of GH84 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2R,3S,4R,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)-1-pentyl-pyrrolidin-2-yl]-N-methyl-ethanamide, CALCIUM ION, ... | | Authors: | Bergeron-Brlek, M, Goodwin-Tindall, J, Cekic, N, Varghese, V, Zandberg, W.F, Shan, X, Roth, C, Chan, S, Davies, G.J, Vocadlo, D.J, Britton, R. | | Deposit date: | 2015-08-05 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Convenient Approach to Stereoisomeric Iminocyclitols: Generation of Potent Brain-Permeable Oga Inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6NZN
 
 | | Dimer-of-dimer amyloid fibril structure of glucagon | | Descriptor: | Glucagon | | Authors: | Gelenter, M.D, Smith, K.J, Liao, S.Y, Mandala, V.S, Dregni, A.J, Lamm, M.S, Tian, Y, Wei, X, Pochan, D.J, Tucker, T.J, Su, Y, Hong, M. | | Deposit date: | 2019-02-14 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The peptide hormone glucagon forms amyloid fibrils with two coexisting beta-strand conformations.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7QZ1
 
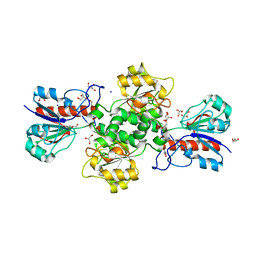 | | Formate dehydrogenase from Starkeya novella | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Formate dehydrogenase, ... | | Authors: | Pontillo, N, Slotboom, D.J, Guskov, A. | | Deposit date: | 2022-01-30 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and structural insight into the chemical resistance and cofactor specificity of the formate dehydrogenase from Starkeya novella.
Febs J., 290, 2023
|
|
3LQH
 
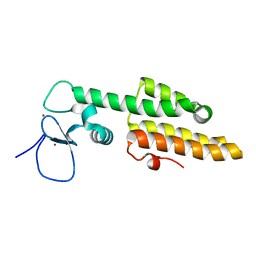 | | Crystal structure of MLL1 PHD3-Bromo in the free form | | Descriptor: | Histone-lysine N-methyltransferase MLL, ZINC ION | | Authors: | Wang, Z, Patel, D.J. | | Deposit date: | 2010-02-09 | | Release date: | 2010-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Pro isomerization in MLL1 PHD3-bromo cassette connects H3K4me readout to CyP33 and HDAC-mediated repression.
Cell(Cambridge,Mass.), 141, 2010
|
|
6O6T
 
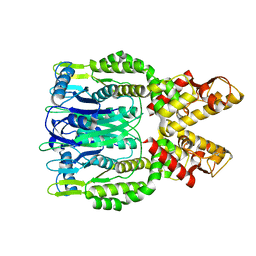 | | Crystal structure of Csm6 H132A mutant | | Descriptor: | Csm6 | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | CRISPR-Cas III-A Csm6 CARF Domain Is a Ring Nuclease Triggering Stepwise cA4Cleavage with ApA>p Formation Terminating RNase Activity.
Mol.Cell, 75, 2019
|
|
6O75
 
 | | Crystal structure of Csm1-Csm4 cassette in complex with pppApA | | Descriptor: | CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), Csm4, MANGANESE (II) ION, ... | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Second Messenger cA4Formation within the Composite Csm1 Palm Pocket of Type III-A CRISPR-Cas Csm Complex and Its Release Path.
Mol.Cell, 75, 2019
|
|
6U9T
 
 | | Wild-type MthK pore in 50 mM K+ | | Descriptor: | Calcium-gated potassium channel MthK, HEXANE-1,6-DIOL, POTASSIUM ION | | Authors: | Posson, D.J, Nimigean, C.M. | | Deposit date: | 2019-09-09 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Selectivity filter ion binding affinity determines inactivation in a potassium channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5BO0
 
 | |
4BZ1
 
 | | Structure of dengue virus EDIII in complex with Fab 3e31 | | Descriptor: | CHLORIDE ION, ENVELOPE PROTEIN, FAB 3E31 HEAVY CHAIN, ... | | Authors: | Li, J, Watterson, D, Chang, C.W, Li, X.Q, Ericsson, D.J, Qiu, L.W, Cai, J.P, Chen, J, Fry, S.R, Cooper, M.A, Che, X.Y, Young, P.R, Kobe, B. | | Deposit date: | 2013-07-23 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Dengue Virus Ediii in Complex with Fab 3E31
To be Published
|
|
4BV0
 
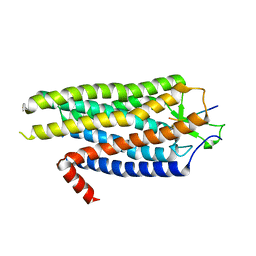 | | High Resolution Structure of Evolved Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion | | Descriptor: | NEUROTENSIN RECEPTOR TYPE 1, NEUROTENSIN/NEUROMEDIN N | | Authors: | Egloff, P, Hillenbrand, M, Scott, D.J, Schlinkmann, K.M, Heine, P, Balada, S, Batyuk, A, Mittl, P, Schuetz, M, Plueckthun, A. | | Deposit date: | 2013-06-24 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7RH6
 
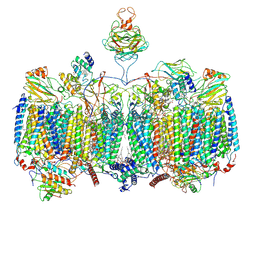 | | Mycobacterial CIII2CIV2 supercomplex, inhibitor free, -Lpqe cyt cc open | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Di Trani, J.M, Yanofsky, D.J, Rubinstein, J.L. | | Deposit date: | 2021-07-16 | | Release date: | 2021-08-04 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of mycobacterial CIII 2 CIV 2 respiratory supercomplex bound to the tuberculosis drug candidate telacebec (Q203).
Elife, 10, 2021
|
|
