7AM4
 
 | |
7AM7
 
 | |
4NHA
 
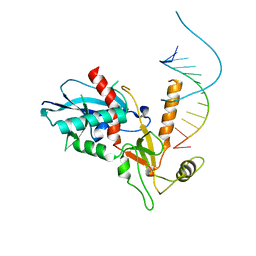
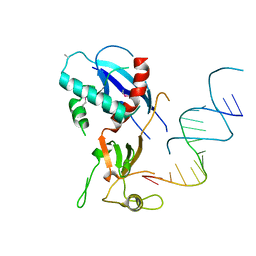 | | Structure of human Dicer Platform-PAZ-Connector Helix cassette in complex with 16-mer siRNA having 5'-p and UU-3' ends (3.4 Angstrom resolution) | | Descriptor: | 5'-R(P*GP*CP*GP*UP*UP*GP*GP*CP*CP*AP*AP*CP*GP*CP*UP*U)-3', Endoribonuclease Dicer | | Authors: | Simanshu, D.K, Tian, Y, Ma, J.-B, Patel, D.J. | | Deposit date: | 2013-11-04 | | Release date: | 2014-03-05 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | A Phosphate-Binding Pocket within the Platform-PAZ-Connector Helix Cassette of Human Dicer.
Mol.Cell, 53, 2014
|
|
4NH5
 
 | |
4NGG
 
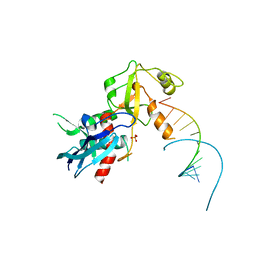 | |
4NGC
 
 | |
4NH6
 
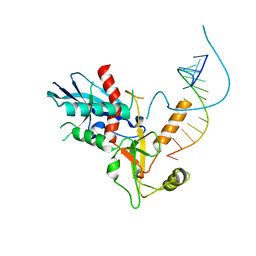 | |
4NTI
 
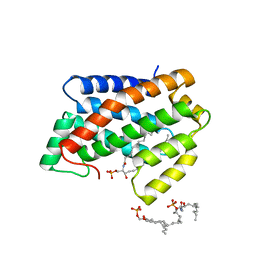 | | Crystal structure of D60N mutant of Arabidopsis ACD11 (accelerated-cell-death 11) complexed with C12 ceramide-1-phosphate (d18:1/12:0) at 2.9 Angstrom resolution | | Descriptor: | (2S,3R,4E)-2-(dodecanoylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, DI(HYDROXYETHYL)ETHER, accelerated-cell-death 11 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-12-02 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Arabidopsis Accelerated Cell Death 11, ACD11, Is a Ceramide-1-Phosphate Transfer Protein and Intermediary Regulator of Phytoceramide Levels.
Cell Rep, 6, 2014
|
|
4O1Z
 
 | | Crystal Structure of Ovine Cyclooxygenase-1 Complex with Meloxicam | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-hydroxy-2-methyl-N-(5-methyl-1,3-thiazol-2-yl)-2H-1,2-benzothiazine-3-carboxamide 1,1-dioxide, ... | | Authors: | Xu, S, Hermanson, D.J, Banerjee, S, Ghebreselasie, K, Clayton, G.M, Garavito, R.M, Marnett, L.J. | | Deposit date: | 2013-12-16 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Oxicams Bind in a Novel Mode to the Cyclooxygenase Active Site via a Two-water-mediated H-bonding Network.
J.Biol.Chem., 289, 2014
|
|
4NTO
 
 | | Crystal structure of D60A mutant of Arabidopsis ACD11 (accelerated-cell-death 11) complexed with C2 ceramide-1-phosphate (d18:1/2:0) at 2.15 Angstrom resolution | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, DI(HYDROXYETHYL)ETHER, accelerated-cell-death 11 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-12-02 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Arabidopsis Accelerated Cell Death 11, ACD11, Is a Ceramide-1-Phosphate Transfer Protein and Intermediary Regulator of Phytoceramide Levels.
Cell Rep, 6, 2014
|
|
4NT1
 
 | | Crystal structure of apo-form of Arabidopsis ACD11 (accelerated-cell-death 11) at 1.8 Angstrom resolution | | Descriptor: | SODIUM ION, accelerated-cell-death 11 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2014-02-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Arabidopsis Accelerated Cell Death 11, ACD11, Is a Ceramide-1-Phosphate Transfer Protein and Intermediary Regulator of Phytoceramide Levels.
Cell Rep, 6, 2014
|
|
8FRR
 
 | | Wild-type myocilin olfactomedin domain | | Descriptor: | CALCIUM ION, GLYCEROL, Myocilin, ... | | Authors: | Ma, M.T, Lieberman, R.L, Huard, D.J.E. | | Deposit date: | 2023-01-08 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Competition between inside-out unfolding and pathogenic aggregation in an amyloid-forming beta-propeller.
Nat Commun, 15, 2024
|
|
4NUF
 
 | | Crystal Structure of SHP/EID1 | | Descriptor: | EID1 peptide, Maltose ABC transporter periplasmic protein, Nuclear receptor subfamily 0 group B member 2 chimeric construct, ... | | Authors: | Zhi, X, Zhou, X.E, He, Y, Zechner, C, Suino-Powell, K.M, Kliewer, S.A, Melcher, K, Mangelsdorf, D.J, Xu, H.E. | | Deposit date: | 2013-12-03 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into gene repression by the orphan nuclear receptor SHP.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7AWK
 
 | |
4P7O
 
 | | Structure of Escherichia coli PgaB C-terminal domain, P1 crystal form | | Descriptor: | Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7B6W
 
 | | Crystal structure of the human alpha1B adrenergic receptor in complex with inverse agonist (+)-cyclazosin | | Descriptor: | Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor, [(4~{a}~{R},8~{a}~{S})-4-(4-azanyl-6,7-dimethoxy-quinazolin-2-yl)-2,3,4~{a},5,6,7,8,8~{a}-octahydroquinoxalin-1-yl]-(furan-2-yl)methanone | | Authors: | Deluigi, M, Morstein, L, Hilge, M, Schuster, M, Merklinger, L, Klipp, A, Scott, D.J, Plueckthun, A. | | Deposit date: | 2020-12-08 | | Release date: | 2022-01-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.873 Å) | | Cite: | Crystal structure of the alpha 1B -adrenergic receptor reveals molecular determinants of selective ligand recognition.
Nat Commun, 13, 2022
|
|
4N2C
 
 | | Crystal structure of Protein Arginine Deiminase 2 (F221/222A, 10 mM Ca2+) | | Descriptor: | CALCIUM ION, Protein-arginine deiminase type-2 | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.022 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
4N2H
 
 | | Crystal structure of Protein Arginine Deiminase 2 (D177A, 0 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Protein-arginine deiminase type-2 | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.808 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
7A98
 
 | | SARS-CoV-2 Spike Glycoprotein with 3 ACE2 Bound | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
4PHM
 
 | | The Structural Basis of Differential Inhibition of Human Calpain by Indole and Phenyl alpha-Mercaptoacrylic Acids | | Descriptor: | 3-(5-bromo-1H-indol-3-yl)-2-thioxopropanoic acid, CALCIUM ION, Calpain small subunit 1 | | Authors: | Rizkallah, P.J, Allemann, R.K, Adams, S.E, Miller, D.J, Hallett, M.B. | | Deposit date: | 2014-05-06 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The structural basis of differential inhibition of human calpain by indole and phenyl alpha-mercaptoacrylic acids.
J.Struct.Biol., 187, 2014
|
|
4POP
 
 | | ThiT with LMG139 bound | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 4-[(4-amino-2-methylpyrimidin-5-yl)methyl]-3-methylthiophene-2-carbaldehyde, ... | | Authors: | Swier, L.J.Y.M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2014-02-26 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Potent Small-Molecule Binders to the S-Component of the ECF Transporter for Thiamine.
Chembiochem, 16, 2015
|
|
4PEH
 
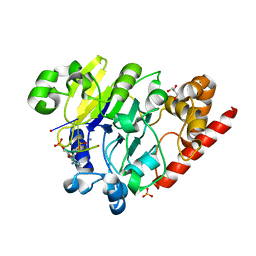 | | Dbr1 in complex with synthetic linear RNA | | Descriptor: | GLYCEROL, MANGANESE (II) ION, RNA (5'-R(*CP*UP*AP*(A2P)P*AP*CP*AP*A)-3'), ... | | Authors: | Montemayor, E.J, Katolik, A, Clark, N.E, Taylor, A.B, Schuermann, J.P, Combs, D.J, Johnsson, R, Holloway, S.P, Stevens, S.W, Damha, M.J, Hart, P.J. | | Deposit date: | 2014-04-23 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of lariat RNA recognition by the intron debranching enzyme Dbr1.
Nucleic Acids Res., 42, 2014
|
|
4P7Q
 
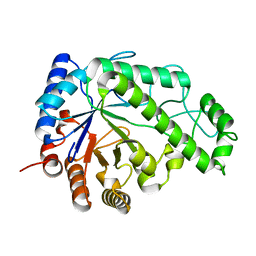 | | Structure of Escherichia coli PgaB C-terminal domain in complex with N-acetylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7BNN
 
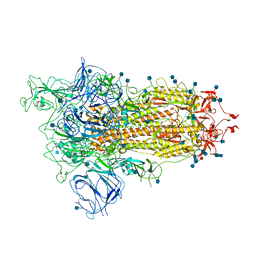 | | Open conformation of D614G SARS-CoV-2 spike with 1 Erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4P31
 
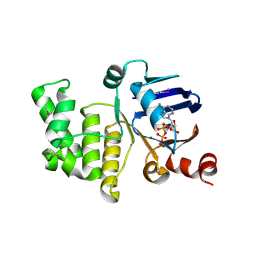 | | Crystal structure of a selenomethionine derivative of E. coli LptB in complex with ADP-Magensium | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lipopolysaccharide export system ATP-binding protein LptB, MAGNESIUM ION | | Authors: | Sherman, D.J, Lazarus, M.B, Murphy, L, Liu, C, Walker, S, Ruiz, N, Kahne, D. | | Deposit date: | 2014-03-05 | | Release date: | 2014-03-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Decoupling catalytic activity from biological function of the ATPase that powers lipopolysaccharide transport.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
