1XYR
 
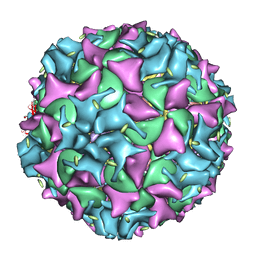 | | Poliovirus 135S cell entry intermediate | | Descriptor: | Genome polyprotein, Coat protein VP1, Coat protein VP2, ... | | Authors: | Bubeck, D, Filman, D.J, Cheng, N, Steven, A.C, Hogle, J.M, Belnap, D.M. | | Deposit date: | 2004-11-10 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | The structure of the poliovirus 135S cell entry intermediate at 10-angstrom resolution reveals the location of an externalized polypeptide that binds to membranes.
J.Virol., 79, 2005
|
|
5DKN
 
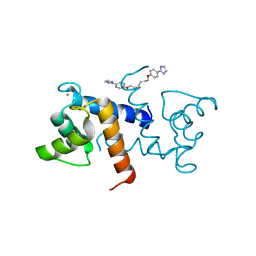 | | Crystal Structure of Calcium-loaded S100B bound to SBi4225 | | Descriptor: | 2,2'-[heptane-1,7-diylbis(oxybenzene-4,1-diyl)]bis(1H-imidazole), CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Ansari, M.I, Pierce, A.D, Wilder, P.T, McKnight, L.E, Raman, E.P, Neau, D.B, Bezawada, P, Alasady, M.J, Varney, K.M, Toth, E.A, MacKerell Jr, A.D, Coop, A, Weber, D.J. | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.528 Å) | | Cite: | Small Molecule Inhibitors of Ca(2+)-S100B Reveal Two Protein Conformations.
J.Med.Chem., 59, 2016
|
|
2YA0
 
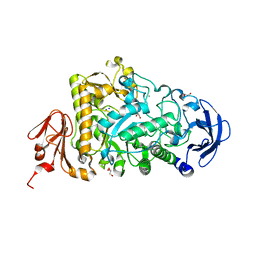 | | Catalytic Module of the Multi-modular glycogen-degrading pneumococcal virulence factor SpuA | | Descriptor: | CALCIUM ION, GLYCEROL, PUTATIVE ALKALINE AMYLOPULLULANASE, ... | | Authors: | Lammerts van Bueren, A, Ficko-Blean, E, Pluvinage, B, Hehemann, J.H, Higgins, M.A, Deng, L, Ogunniyi, A.D, Stroeher, U.H, Warry, N.E, Burke, R.D, Czjzek, M, Paton, J.C, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-02-17 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Conformation and Function of a Multimodular Glycogen-Degrading Pneumococcal Virulence Factor.
Structure, 19, 2011
|
|
1XU9
 
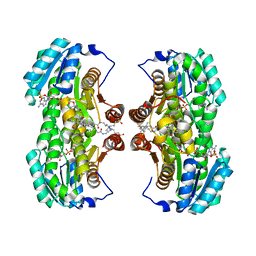 | | Crystal Structure of the Interface Closed Conformation of 11b-hydroxysteroid dehydrogenase isozyme 1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Corticosteroid 11-beta-dehydrogenase, ... | | Authors: | Hosfield, D.J, Wu, Y, Skene, R.J, Hilger, M, Jennings, A, Snell, G.P, Aertgeerts, K. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Conformational Flexibility in Crystal Structures of Human 11beta-hydroxysteroid dehydrogenase type I provide insights into glucocorticoid interconversion and enzyme regulation.
J.Biol.Chem., 280, 2005
|
|
6U9T
 
 | | Wild-type MthK pore in 50 mM K+ | | Descriptor: | Calcium-gated potassium channel MthK, HEXANE-1,6-DIOL, POTASSIUM ION | | Authors: | Posson, D.J, Nimigean, C.M. | | Deposit date: | 2019-09-09 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Selectivity filter ion binding affinity determines inactivation in a potassium channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1XGB
 
 | |
1XGC
 
 | |
8BBZ
 
 | |
6RH5
 
 | | Solution structure and 1H, 13C and 15N chemical shift assignments for NECAP1 PHear domain | | Descriptor: | Adaptin ear-binding coat-associated protein 1 | | Authors: | Owen, D.J, Neuhaus, D, Yang, J.-C, Herrmann, T. | | Deposit date: | 2019-04-18 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Dev.Cell, 50, 2019
|
|
8BHU
 
 | | Crystal Structure of SilF (Ag(I) form | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SILVER ION, SULFATE ION, ... | | Authors: | Lithgo, R.M, Carr, S.B, Quigley, A.M, Scott, D.J. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of SilF (apo form)
To Be Published
|
|
1UOV
 
 | | Calcium binding domain C2B | | Descriptor: | CALCIUM ION, GLYCEROL, SYNAPTOTAGMIN I | | Authors: | Cheng, Y, Sequeira, S.M, Sollner, T.H, Patel, D.J. | | Deposit date: | 2003-09-24 | | Release date: | 2004-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallographic Identification of Ca2+ and Sr2+ Coordination Sites in Synaptotagmin I C2B Domain
Protein Sci., 13, 2004
|
|
5DOB
 
 | | Crystal structure of the Human Cytomegalovirus Nuclear Egress Complex (NEC) | | Descriptor: | CALCIUM ION, Virion egress protein UL31 homolog, Virion egress protein UL34 homolog, ... | | Authors: | Lye, M.F, El Omari, K, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Unexpected features and mechanism of heterodimer formation of a herpesvirus nuclear egress complex.
Embo J., 34, 2015
|
|
2B5W
 
 | | Crystal structure of D38C glucose dehydrogenase mutant from Haloferax mediterranei | | Descriptor: | CITRATE ANION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, ... | | Authors: | Britton, K.L, Baker, P.J, Fisher, M, Ruzheinikov, S, Gilmour, D.J, Bonete, M.-J, Ferrer, J, Pire, C, Esclapez, J, Rice, D.W. | | Deposit date: | 2005-09-29 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Analysis of protein solvent interactions in glucose dehydrogenase from the extreme halophile Haloferax mediterranei.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6TQE
 
 | | The structure of ABC transporter Rv1819c without addition of substrate | | Descriptor: | ABC transporter ATP-binding protein/permease, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Rempel, S, Gati, C, Slotboom, D.J, Guskov, A. | | Deposit date: | 2019-12-16 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | A mycobacterial ABC transporter mediates the uptake of hydrophilic compounds.
Nature, 580, 2020
|
|
3IPS
 
 | | X-ray structure of benzisoxazole synthetic agonist bound to the LXR-alpha | | Descriptor: | Nuclear receptor coactivator 1, Oxysterols receptor LXR-alpha, SULFATE ION, ... | | Authors: | Fradera, X, Vu, D, Nimz, O, Skene, R, Hosfield, D, Wijnands, R, Cooke, A.J, Haunso, A, King, A, Bennet, D.J, McGuire, R, Uitdehaag, J.C.M. | | Deposit date: | 2009-08-18 | | Release date: | 2010-06-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | X-ray structures of the LXRalpha LBD in its homodimeric form and implications for heterodimer signaling.
J.Mol.Biol., 399, 2010
|
|
2Y37
 
 | | The discovery of novel, potent and highly selective inhibitors of inducible nitric oxide synthase (iNOS) | | Descriptor: | 2-[(1R)-3-amino-1-phenyl-propoxy]-4-chloro-benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, GLYCEROL, ... | | Authors: | Cheshire, D.R, Andrews, G, Beaton, H.G, Birkinshaw, T.N, Boughton-Smith, N, Connolly, S, Cook, T.R, Cooper, A, Cooper, S.L, Cox, D, Dixon, J, Gensmantel, N, Hamley, P.J, Harrison, R, Hartopp, P, Kack, H, Luker, T, Mete, A, Millichip, I, Nicholls, D.J, Pimm, A.D, St-Gallay, S.A, Wallace, A.V. | | Deposit date: | 2010-12-19 | | Release date: | 2011-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Discovery of Novel, Potent and Highly Selective Inhibitors of Inducible Nitric Oxide Synthase (Inos).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2C8I
 
 | | Complex Of Echovirus Type 12 With Domains 1, 2, 3 and 4 Of Its Receptor Decay Accelerating Factor (Cd55) By Cryo Electron Microscopy At 16 A | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR, ECHOVIRUS 11 COAT PROTEIN VP1, ECHOVIRUS 11 COAT PROTEIN VP2, ... | | Authors: | Pettigrew, D.M, Williams, D.T, Kerrigan, D, Evans, D.J, Lea, S.M, Bhella, D. | | Deposit date: | 2005-12-05 | | Release date: | 2006-01-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Structural and Functional Insights Into the Interaction of Echoviruses and Decay-Accelerating Factor.
J.Biol.Chem., 281, 2006
|
|
1XOK
 
 | | crystal structure of alfalfa mosaic virus RNA 3'UTR in complex with coat protein N terminal peptide | | Descriptor: | BROMIDE ION, Coat protein, alfalfa mosaic virus RNA 3' UTR | | Authors: | Guogas, L.M, Filman, D.J, Hogle, J.M, Gehrke, L. | | Deposit date: | 2004-10-06 | | Release date: | 2005-01-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cofolding organizes alfalfa mosaic virus RNA and coat protein for replication.
Science, 306, 2004
|
|
6THN
 
 | | Multiple Genomic RNA-Coat Protein Contacts Play Vital Roles in the Assembly of Infectious Enterovirus-E symmetry expansion+2fold focused classification | | Descriptor: | Genome polyprotein, MYRISTIC ACID, RNA Peak 9 Bernoulli Plot, ... | | Authors: | Chandler-Bostock, R, Mata, C.P, Bingham, R, Dykeman, E.J, Meng, B, Tuthill, T.J, Rowlands, D.J, Ranson, N.A, Twarock, R, Stockley, P.G. | | Deposit date: | 2019-11-20 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Assembly of infectious enteroviruses depends on multiple, conserved genomic RNA-coat protein contacts.
Plos Pathog., 16, 2020
|
|
2YNN
 
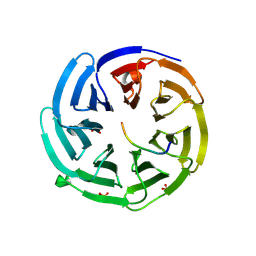 | | yeast betaprime COP 1-304 with KTKTN motif | | Descriptor: | COATOMER SUBUNIT BETA', KTKTN MOTIF, SULFATE ION | | Authors: | Jackson, L.P, Lewis, M, Kent, H.M, Edeling, M.A, Evans, P.R, Duden, R, Owen, D.J. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | Molecular Basis for Recognition of Dilysine Trafficking Motifs by Copi.
Dev.Cell, 23, 2012
|
|
2NAV
 
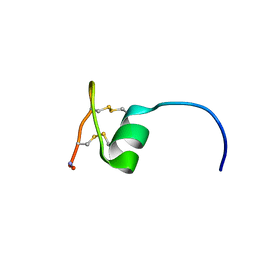 | | NMR solution structure of Ex-4[1-16]/pl14a | | Descriptor: | Exendin-4, Alpha/kappa-conotoxin pl14a chimera | | Authors: | Schroeder, C.I, Swedberg, J.E, Craik, D.J. | | Deposit date: | 2016-01-11 | | Release date: | 2016-05-04 | | Last modified: | 2016-08-17 | | Method: | SOLUTION NMR | | Cite: | Truncated Glucagon-like Peptide-1 and Exendin-4 alpha-Conotoxin pl14a Peptide Chimeras Maintain Potency and alpha-Helicity and Reveal Interactions Vital for cAMP Signaling in Vitro.
J.Biol.Chem., 291, 2016
|
|
2NAW
 
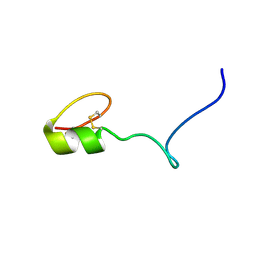 | |
2B5V
 
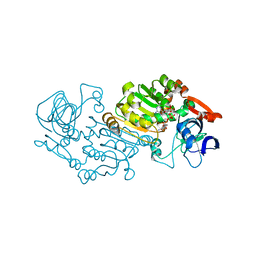 | | Crystal structure of glucose dehydrogenase from Haloferax mediterranei | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, glucose dehydrogenase | | Authors: | Britton, K.L, Baker, P.J, Fisher, M, Ruzheinikov, S, Gilmour, D.J, Bonete, M.-J, Ferrer, J, Pire, C, Esclapez, J, Rice, D.W. | | Deposit date: | 2005-09-29 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of protein solvent interactions in glucose dehydrogenase from the extreme halophile Haloferax mediterranei.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
7UDQ
 
 | | Crystal structure of COQ8A-CA157 inhibitor complex in space group P1 | | Descriptor: | 4-[(3,4,5-trimethoxyphenyl)amino]quinoline-7-carbonitrile, Atypical kinase COQ8A, mitochondrial, ... | | Authors: | Bingman, C.A, Murray, N, Smith, R.W, Pagliarini, D.J. | | Deposit date: | 2022-03-20 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small-molecule inhibition of the archetypal UbiB protein COQ8.
Nat.Chem.Biol., 19, 2023
|
|
7UDP
 
 | | Crystal structure of COQ8A-CA157 inhibitor complex in space group C2 | | Descriptor: | 4-[(3,4,5-trimethoxyphenyl)amino]quinoline-7-carbonitrile, Atypical kinase COQ8A, mitochondrial | | Authors: | Bingman, C.A, Murray, N, Smith, R.W, Pagliarini, D.J. | | Deposit date: | 2022-03-20 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Small-molecule inhibition of the archetypal UbiB protein COQ8.
Nat.Chem.Biol., 19, 2023
|
|
