4EB1
 
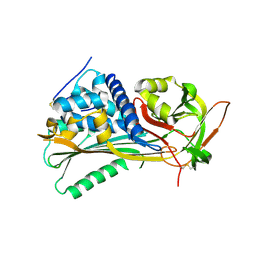 | | Hyperstable in-frame insertion variant of antithrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antithrombin-III | | Authors: | Martinez-Martinez, I, Johnson, D.J.D, Yamasaki, M, Corral, J, Huntington, J.A. | | Deposit date: | 2012-03-23 | | Release date: | 2012-07-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Type II antithrombin deficiency caused by a large in-frame insertion: structural, functional and pathological relevance.
J.Thromb.Haemost., 10, 2012
|
|
1NYG
 
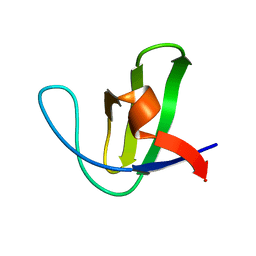 | |
5TA8
 
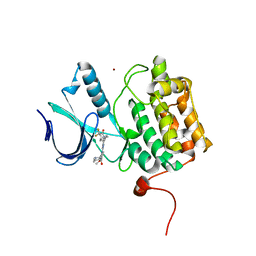 | | Crystal structure of PLK1 in complex with a novel 5,6-dihydroimidazolo[1,5-f]pteridine inhibitor | | Descriptor: | 4-[(9-cyclopentyl-7,7-difluoro-5-methyl-6-oxo-6,7,8,9-tetrahydro-5H-pyrimido[4,5-b][1,4]diazepin-2-yl)amino]-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Serine/threonine-protein kinase PLK1, ZINC ION | | Authors: | Skene, R.J, Hosfield, D.J. | | Deposit date: | 2016-09-09 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design and SAR development of 5,6-dihydroimidazolo[1,5-f]pteridine derivatives as novel Polo-like kinase-1 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5TA6
 
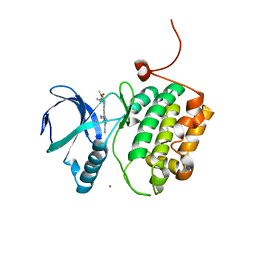 | | Crystal structure of PLK1 in complex with a novel 5,6-dihydroimidazolo[1,5-f]pteridine inhibitor. | | Descriptor: | 4-{[(6R)-7-cyano-5-cyclopentyl-6-ethyl-5,6-dihydroimidazo[1,5-f]pteridin-3-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Serine/threonine-protein kinase PLK1, ZINC ION | | Authors: | Skene, R.J, Hosfield, D.J. | | Deposit date: | 2016-09-09 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based design and SAR development of 5,6-dihydroimidazolo[1,5-f]pteridine derivatives as novel Polo-like kinase-1 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
9D3G
 
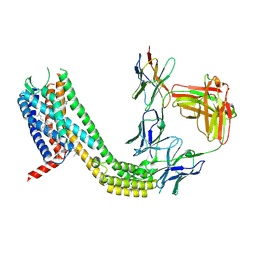 | | Cryo-EM structure of CCR6 bound by SQA1 and OXM1 | | Descriptor: | 1-(4-chlorophenyl)-N-{[(2R)-4-(2,3-dihydro-1H-inden-2-yl)-5-oxomorpholin-2-yl]methyl}cyclopropane-1-carboxamide, 4-[[3,4-bis(oxidanylidene)-2-[[(1~{R})-1-(4-propan-2-ylfuran-2-yl)propyl]amino]cyclobuten-1-yl]amino]-~{N},~{N}-dimethyl-3-oxidanyl-pyridine-2-carboxamide, CCR6, ... | | Authors: | Wasilko, D.J, Wu, H. | | Deposit date: | 2024-08-10 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis for CCR6 modulation by allosteric antagonists.
Nat Commun, 15, 2024
|
|
8ZFO
 
 | |
9G9Q
 
 | | Crystal structure of PbdA bound to p-methoxybenzoate. | | Descriptor: | 4-METHOXYBENZOIC ACID, Cytochrome P450 CYP199, GLYCEROL, ... | | Authors: | Hinchen, D.J, Wolf, M.E, Eltis, L.D, McGeehan, J.E. | | Deposit date: | 2024-07-25 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Characterization of a cytochrome P450 that catalyzes the O-demethylation of lignin-derived benzoates.
J.Biol.Chem., 300, 2024
|
|
9DTT
 
 | |
8ZFR
 
 | |
9C67
 
 | |
9CSI
 
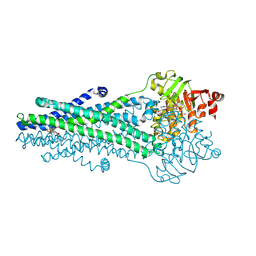 | | A. baumannii MsbA Bound to Cerastecin Compound 5 | | Descriptor: | 3,3'-[(1,4-dioxobutane-1,4-diyl)bis(azanediyl)]bis[(4-butylbenzene-1-sulfonamido)benzoic acid], Lipid A export ATP-binding/permease protein MsbA, MAGNESIUM ION, ... | | Authors: | Klein, D.J, Ishchenko, A, Soisson, S, Cheng, R, Hennig, M. | | Deposit date: | 2024-07-23 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cerastecin Inhibition of the Lipooligosaccharide Transporter MsbA to Combat Acinetobacter baumannii : From Screening Impurity to In Vivo Efficacy.
J.Med.Chem., 67, 2024
|
|
8ZFP
 
 | |
9AU7
 
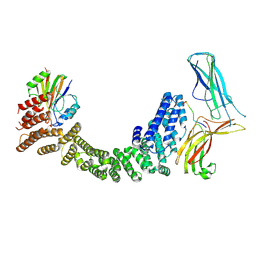 | | Human Retriever VPS35L/VPS29/VPS26C complex bound to SNX17 peptide (Composite Map) | | Descriptor: | Sorting nexin-17, VPS35 endosomal protein-sorting factor-like, Vacuolar protein sorting-associated protein 26C, ... | | Authors: | Chen, B, Chen, Z, Han, Y, Boesch, D.J, Juneja, P, Burstein, E, Fung, H.Y.J. | | Deposit date: | 2024-02-28 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for Retriever-SNX17 assembly and endosomal sorting
To be published
|
|
9BEQ
 
 | | Crystal structure of pregnane X receptor ligand binding domain complexed with AP1867 | | Descriptor: | Nuclear receptor subfamily 1 group I member 2, {3-[3-(3,4-DIMETHOXY-PHENYL)-1-(1-{1-[2-(3,4,5-TRIMETHOXY-PHENYL)-BUTYRYL]-PIPERIDIN-2YL}-VINYLOXY)-PROPYL]-PHENOXY}-ACETIC ACID | | Authors: | Huber, A.D, Poudel, S, Miller, D.J, Chen, T. | | Deposit date: | 2024-04-16 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | PROTAC-mediated activation, rather than degradation, of a nuclear receptor reveals complex ligand-receptor interaction network.
Structure, 2024
|
|
9CSG
 
 | | Human Serum Albumin Bound to Cerastecin Compound 5e | | Descriptor: | 2-(4-butylbenzene-1-sulfonamido)-5-(4-{3-carboxy-4-[4-(2-methoxyethyl)benzene-1-sulfonamido]anilino}-4-oxobutanamido)benzoic acid, Albumin, MYRISTIC ACID | | Authors: | Hruza, A, Klein, D.J, Ishchenko, A. | | Deposit date: | 2024-07-23 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | Cerastecin Inhibition of the Lipooligosaccharide Transporter MsbA to Combat Acinetobacter baumannii : From Screening Impurity to In Vivo Efficacy.
J.Med.Chem., 67, 2024
|
|
9GS9
 
 | | Tn7016 PseCAST QCascade | | Descriptor: | Cas6, Cas7.1, Cas8, ... | | Authors: | Lampe, G.D, Liang, A.R, Zhang, D.J, Fernandez, I.S, Sternberg, S.H. | | Deposit date: | 2024-09-13 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure-guided engineering of type I-F CASTs for targeted gene insertion in human cells.
Biorxiv, 2024
|
|
8ZFT
 
 | |
9HVP
 
 | | Design, activity and 2.8 Angstroms crystal structure of a C2 symmetric inhibitor complexed to HIV-1 protease | | Descriptor: | HIV-1 Protease, benzyl [(1R,4S,6S,9R)-4,6-dibenzyl-5-hydroxy-1,9-bis(1-methylethyl)-2,8,11-trioxo-13-phenyl-12-oxa-3,7,10-triazatridec-1-yl]carbamate | | Authors: | Neidhart, D.J, Erickson, J. | | Deposit date: | 1990-11-06 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design, activity, and 2.8 A crystal structure of a C2 symmetric inhibitor complexed to HIV-1 protease.
Science, 249, 1990
|
|
8ZFN
 
 | |
9C77
 
 | | cryoEM structure of CRISPR associated effector, CARF-Adenosine deaminase 1, Cad1, in cA4 bound form with ATP. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenosine deaminase domain-containing protein, MAGNESIUM ION, ... | | Authors: | Majumder, P, Patel, D.J. | | Deposit date: | 2024-06-10 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The CRISPR-associated adenosine deaminase Cad1 converts ATP to ITP to provide antiviral immunity.
Cell, 2024
|
|
8ZFQ
 
 | |
9CKJ
 
 | | Co-crystal structure of 53BP1 tandem Tudor domains in complex with UNC9512 | | Descriptor: | (3S)-1-[4-(4-methylpiperazin-1-yl)pyridine-2-carbonyl]-N-[(3M)-3-(3-oxo-2,3-dihydro-1H-isoindol-5-yl)phenyl]piperidine-3-carboxamide, TP53-binding protein 1, UNKNOWN ATOM OR ION | | Authors: | Zeng, H, Dong, A, Shell, D.J, Foley, C, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-07-09 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Co-crystal structure of 53BP1 tandem Tudor domains in complex with UNC9512
To be published
|
|
8ZFS
 
 | |
9C8S
 
 | |
9G9S
 
 | | Crystal structure of PbdA bound to veratrate | | Descriptor: | 3,4-dimethoxybenzoic acid, Cytochrome P450 CYP199, GLYCEROL, ... | | Authors: | Hinchen, D.J, Wolf, M.E, Eltis, L.D, McGeehan, J.E. | | Deposit date: | 2024-07-25 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Characterization of a cytochrome P450 that catalyzes the O-demethylation of lignin-derived benzoates.
J.Biol.Chem., 300, 2024
|
|
