3HXQ
 
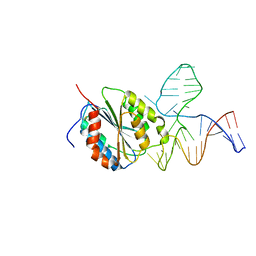 | | Crystal Structure of Von Willebrand Factor (VWF) A1 Domain in Complex with DNA Aptamer ARC1172, an Inhibitor of VWF-Platelet Binding | | Descriptor: | Aptamer ARC1172, von Willebrand Factor | | Authors: | Huang, R.H, Sadler, J.E, Fremont, D.H, Diener, J.L, Schaub, R.G. | | Deposit date: | 2009-06-21 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | A structural explanation for the antithrombotic activity of ARC1172, a DNA aptamer that binds von Willebrand factor domain A1.
Structure, 17, 2009
|
|
5W7M
 
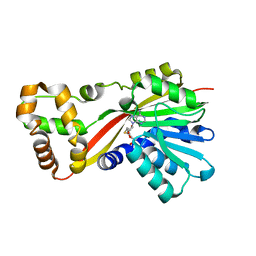 | | Crystal structure of RoqN | | Descriptor: | Glandicoline B O-methyltransferase roqN, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Newmister, S.A, Romminger, S, Schmidt, J.J, Williams, R.M, Smith, J.L, Berlinck, R.G.S, Sherman, D.H. | | Deposit date: | 2017-06-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Unveiling sequential late-stage methyltransferase reactions in the meleagrin/oxaline biosynthetic pathway.
Org. Biomol. Chem., 16, 2018
|
|
5WEU
 
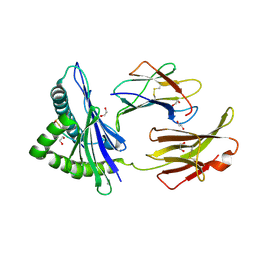 | | Crystal Structure of H2-Dd with disulfide-linked 10mer peptide | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Envelope glycoprotein gp160, ... | | Authors: | Jiang, J.S, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2017-07-10 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.584 Å) | | Cite: | Crystal structure of a TAPBPR-MHC I complex reveals the mechanism of peptide editing in antigen presentation.
Science, 358, 2017
|
|
5WES
 
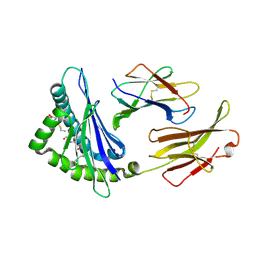 | | Crystal Structure H2-Dd with disulfide-linked 5mer peptide | | Descriptor: | Beta-2-microglobulin, GLYCINE, H-2 class I histocompatibility antigen, ... | | Authors: | Jiang, J.S, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2017-07-10 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | Crystal structure of a TAPBPR-MHC I complex reveals the mechanism of peptide editing in antigen presentation.
Science, 358, 2017
|
|
5UUC
 
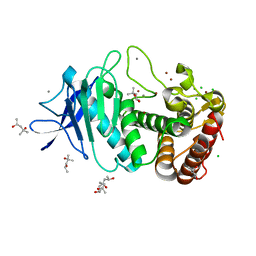 | | Tetragonal thermolysin cryocooled to 100 K with 50% mpd as cryoprotectant | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Juers, D.H. | | Deposit date: | 2017-02-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.60000932 Å) | | Cite: | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4P85
 
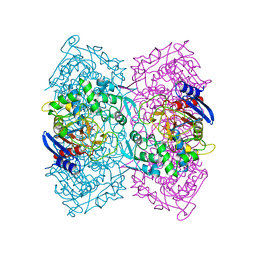 | | Crystal structure of Est-Y29, a novel penicillin-binding protein/beta-lactamase homolog from a metagenomic library | | Descriptor: | DIETHYL PHOSPHONATE, Est-Y29 | | Authors: | Ngo, T.D, Ryu, B.H, Ju, H.S, Jang, E.J, Kim, K.K, Kim, D.H. | | Deposit date: | 2014-03-30 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic analysis and biochemical applications of a novel penicillin-binding protein/ beta-lactamase homologue from a metagenomic library.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2G3S
 
 | | RNA structure containing GU base pairs | | Descriptor: | 5'-R(*GP*GP*CP*GP*UP*GP*CP*C)-3', MAGNESIUM ION | | Authors: | Jang, S.B, Hung, L.W, Jeong, M.S, Holbrook, E.L, Chen, X, Turner, D.H, Holbrook, S.R. | | Deposit date: | 2006-02-20 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | The crystal structure at 1.5 angstroms resolution of an RNA octamer duplex containing tandem G.U basepairs
Biophys.J., 90, 2006
|
|
4OR5
 
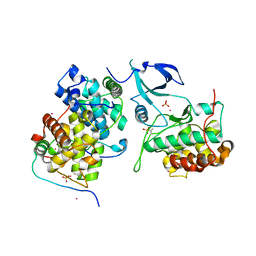 | | Crystal structure of HIV-1 Tat complexed with human P-TEFb and AFF4 | | Descriptor: | AF4/FMR2 family member 4, Cyclin-T1, Cyclin-dependent kinase 9, ... | | Authors: | Gu, J, Babayeva, N.D, Suwa, Y, Baranovskiy, A.G, Price, D.H, Tahirov, T.H. | | Deposit date: | 2014-02-10 | | Release date: | 2014-04-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of HIV-1 Tat complexed with human P-TEFb and AFF4.
Cell Cycle, 13, 2014
|
|
5UU7
 
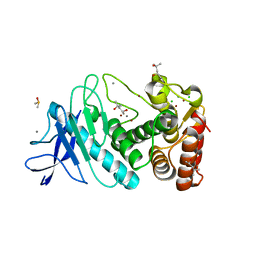 | | Tetragonal thermolysin (295 K) in the presence of 50% mpd | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Juers, D.H. | | Deposit date: | 2017-02-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.60001385 Å) | | Cite: | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5UUE
 
 | |
4P87
 
 | | Crystal structure of Est-Y29, a novel penicillin-binding protein/beta-lactamase homolog from a metagenomic library | | Descriptor: | 4-NITROPHENYL PHOSPHATE, Est-Y29 | | Authors: | Ngo, T.D, Ryu, B.H, Ju, H.S, Jang, E.J, Kim, K.K, Kim, D.H. | | Deposit date: | 2014-03-30 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Crystallographic analysis and biochemical applications of a novel penicillin-binding protein/ beta-lactamase homologue from a metagenomic library.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5W7R
 
 | | Crystal structure of OxaC in complex with SAH and oxaline | | Descriptor: | (3E,7aR,12aS)-3-[(1H-imidazol-4-yl)methylidene]-6,12-dimethoxy-7a-(2-methylbut-3-en-2-yl)-7a,12-dihydro-1H,5H-imidazo[1 ',2':1,2]pyrido[2,3-b]indole-2,5(3H)-dione, OxaC, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Newmister, S.A, Romminger, S, Schmidt, J.J, Williams, R.M, Smith, J.L, Berlinck, R.G.S, Sherman, D.H. | | Deposit date: | 2017-06-20 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Unveiling sequential late-stage methyltransferase reactions in the meleagrin/oxaline biosynthetic pathway.
Org. Biomol. Chem., 16, 2018
|
|
5WER
 
 | | Crystal Structure of TAPBPR and H2-Dd complex | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CITRIC ACID, ... | | Authors: | Jiang, J.S, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2017-07-10 | | Release date: | 2017-10-18 | | Last modified: | 2019-08-28 | | Method: | X-RAY DIFFRACTION (3.412 Å) | | Cite: | Crystal structure of a TAPBPR-MHC I complex reveals the mechanism of peptide editing in antigen presentation.
Science, 358, 2017
|
|
5WPU
 
 | | Crystal structure HpiC1 Y101S | | Descriptor: | 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
2GRM
 
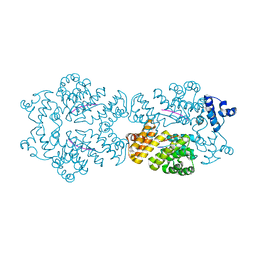 | | Crystal structure of PrgX/iCF10 complex | | Descriptor: | PrgX, peptide | | Authors: | Shi, K, Kozlowicz, B.K, Gu, Z.Y, Ohlendorf, D.H, Earhart, C.A, Dunny, G.M. | | Deposit date: | 2006-04-24 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis for control of conjugation by bacterial pheromone and inhibitor peptides.
Mol.Microbiol., 62, 2006
|
|
2H7X
 
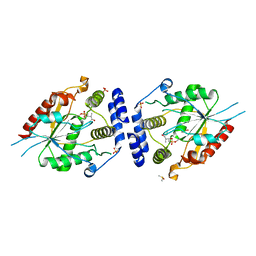 | | Pikromycin Thioesterase adduct with reduced triketide affinity label | | Descriptor: | DIMETHYL SULFOXIDE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Giraldes, J.W, Akey, D.L, Kittendorf, J.D, Sherman, D.H, Smith, J.S, Fecik, R.A. | | Deposit date: | 2006-06-05 | | Release date: | 2006-09-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Mechanistic Insights of Polyketide Macrolactonization from Polyketide-based Affinity Labels
NAT.CHEM.BIOL., 2, 2006
|
|
3HL6
 
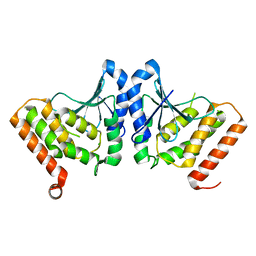 | | Staphylococcus aureus pathogenicity island 3 ORF9 protein | | Descriptor: | CHLORIDE ION, Pathogenicity island protein | | Authors: | Kruse, A.C, Huseby, M, Shi, K, Digre, J, Schlievert, P.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2009-05-26 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional studies of a pathogenicity island protein from Staphylococcus aureus
To be Published
|
|
2H7Y
 
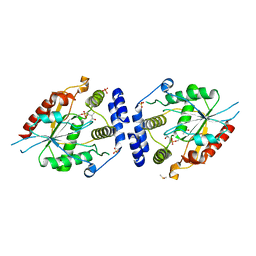 | | Pikromycin Thioesterase with covalent affinity label | | Descriptor: | DIMETHYL SULFOXIDE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Giraldes, J.W, Akey, D.L, Kittendorf, J.D, Sherman, D.H, Smith, J.S, Fecik, R.A. | | Deposit date: | 2006-06-06 | | Release date: | 2006-09-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mechanistic Insights of Polyketide Macrolactonization from Polyketide-based Affinity Labels
NAT.CHEM.BIOL., 2, 2006
|
|
3I0O
 
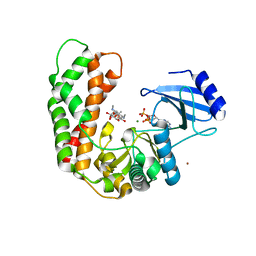 | | Crystal Structure of Spectinomycin Phosphotransferase, APH(9)-Ia, in complex with ADP and Spectinomcyin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NICKEL (II) ION, ... | | Authors: | Fong, D.H, Lemke, C.T, Hwang, J, Xiong, B, Berghuis, A.M. | | Deposit date: | 2009-06-25 | | Release date: | 2010-01-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the antibiotic resistance factor spectinomycin phosphotransferase from Legionella pneumophila.
J.Biol.Chem., 285, 2010
|
|
2HFJ
 
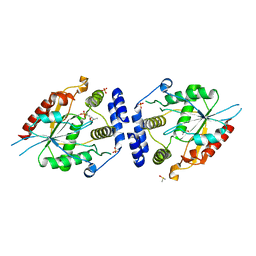 | | Pikromycin thioesterase with covalent pentaketide affinity label | | Descriptor: | DIMETHYL SULFOXIDE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Akey, D.L, Kittendorf, J.D, Giraldes, J.W, Fecik, R.A, Sherman, D.H, Smith, J.L. | | Deposit date: | 2006-06-24 | | Release date: | 2006-09-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for Macrolactonization by the Pikromycin Thioesterase
NAT.CHEM.BIOL., 2, 2006
|
|
5W1U
 
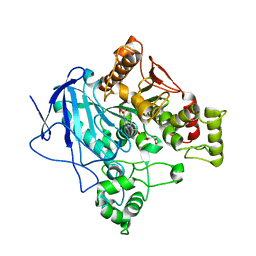 | | Culex quinquefasciatus carboxylesterase B2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Carboxylic ester hydrolase, MALONATE ION | | Authors: | Hopkins, D.H, Jackson, C.J. | | Deposit date: | 2017-06-04 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of an Insecticide Sequestering Carboxylesterase from the Disease Vector Culex quinquefasciatus: What Makes an Enzyme a Good Insecticide Sponge?
Biochemistry, 56, 2017
|
|
4N0A
 
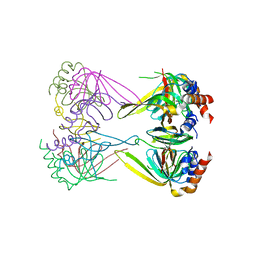 | | Crystal structure of Lsm2-3-Pat1C complex from Saccharomyces cerevisiae | | Descriptor: | DNA topoisomerase 2-associated protein PAT1, U6 snRNA-associated Sm-like protein LSm2, U6 snRNA-associated Sm-like protein LSm3 | | Authors: | Wu, D.H. | | Deposit date: | 2013-10-01 | | Release date: | 2013-12-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Lsm2 and Lsm3 bridge the interaction of the Lsm1-7 complex with Pat1 for decapping activation
Cell Res., 24, 2014
|
|
5WPS
 
 | | Crystal structure HpiC1 Y101F | | Descriptor: | 1,2-ETHANEDIOL, 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.389 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
5WET
 
 | | Crystal Structure of H2-Dd with disulfide-linked 6mer peptide | | Descriptor: | Beta-2-microglobulin, GLYCINE, H-2 class I histocompatibility antigen, ... | | Authors: | Jiang, J.S, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2017-07-10 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of a TAPBPR-MHC I complex reveals the mechanism of peptide editing in antigen presentation.
Science, 358, 2017
|
|
5WPR
 
 | | Crystal structure HpiC1 in C2 space group | | Descriptor: | 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
