6PVJ
 
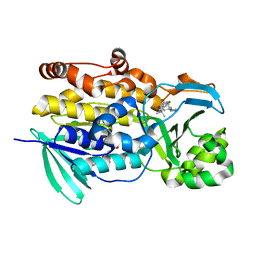 | | Crystal structure of PhqK in complex with malbrancheamide C | | Descriptor: | (5aS,12aS,13aS)-9-bromo-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
3K55
 
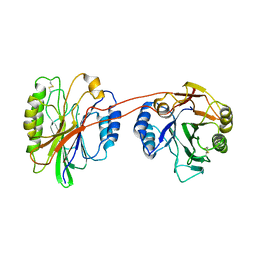 | | Structure of beta hairpin deletion mutant of beta toxin from Staphylococcus aureus | | Descriptor: | Beta-hemolysin, CHLORIDE ION, SODIUM ION | | Authors: | Kruse, A.C, Huseby, M, Shi, K, Digre, J, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2009-10-06 | | Release date: | 2011-01-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structure of a mutant beta toxin from Staphylococcus aureus reveals domain swapping and conformational flexibility
Acta Crystallogr.,Sect.F, 67, 2011
|
|
5J8H
 
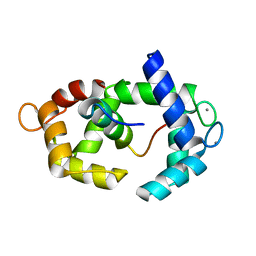 | | Structure of calmodulin in a complex with a peptide derived from a calmodulin-dependent kinase | | Descriptor: | CALCIUM ION, Calmodulin, Eukaryotic elongation factor 2 kinase | | Authors: | Alphonse, S, Lee, K, Piserchio, A, Tavares, C.D.J, Giles, D.H, Wellmann, R.M, Dalby, K.N, Ghose, R. | | Deposit date: | 2016-04-07 | | Release date: | 2016-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Recognition of Eukaryotic Elongation Factor 2 Kinase by Calmodulin.
Structure, 24, 2016
|
|
6ORT
 
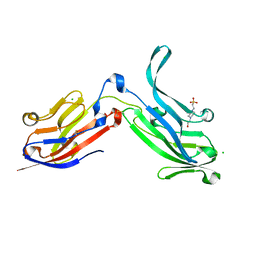 | | Crystal Structure of Bos taurus Mxra8 Ectodomain | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Matrix remodeling-associated protein 8 | | Authors: | Fremont, D.H, Kim, A.S, Nelson, C.A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-04-30 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Evolutionary Insertion in the Mxra8 Receptor-Binding Site Confers Resistance to Alphavirus Infection and Pathogenesis.
Cell Host Microbe, 27, 2020
|
|
7NL3
 
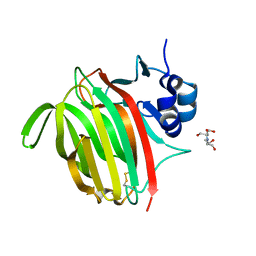 | |
7NDE
 
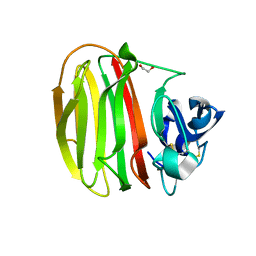 | | Trichoderma parareesei PL7A beta-glucuronan lyase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glucuronan lyase | | Authors: | Fredslund, F, Welner, D.H, Wilkens, C. | | Deposit date: | 2021-02-01 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Glucuronan lyases from family PL7 use a Tyr/Tyr syn beta-elimination catalytic mechanism for glucuronan breakdown.
Chem.Commun.(Camb.), 60, 2024
|
|
7NCZ
 
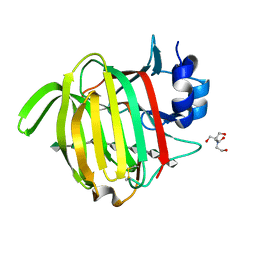 | |
7NM6
 
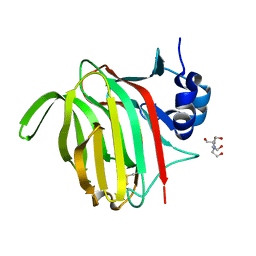 | |
3MI9
 
 | | Crystal structure of HIV-1 Tat complexed with human P-TEFb | | Descriptor: | Cell division protein kinase 9, Cyclin-T1, Protein Tat, ... | | Authors: | Tahirov, T.H, Babayeva, N.D, Varzavand, K, Cooper, J.J, Sedore, S.C, Price, D.H. | | Deposit date: | 2010-04-09 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of HIV-1 Tat complexed with human P-TEFb.
Nature, 465, 2010
|
|
7ZER
 
 | | Crystal structure of UGT85B1 from Sorghum bicolor in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, Cyanohydrin beta-glucosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Putkaradze, N, Fredslund, F, Welner, D.H. | | Deposit date: | 2022-03-31 | | Release date: | 2022-07-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure-guided engineering of key amino acids in UGT85B1 controlling substrate and stereo-specificity in aromatic cyanogenic glucoside biosynthesis.
Plant J., 111, 2022
|
|
7ZF0
 
 | | Crystal structure of UGT85B1 from Sorghum bicolor in complex with UDP and p-hydroxymandelonitrile | | Descriptor: | (2S)-HYDROXY(4-HYDROXYPHENYL)ETHANENITRILE, 1,2-ETHANEDIOL, Cyanohydrin beta-glucosyltransferase, ... | | Authors: | Putkaradze, N, Fredslund, F, Welner, D.H. | | Deposit date: | 2022-03-31 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided engineering of key amino acids in UGT85B1 controlling substrate and stereo-specificity in aromatic cyanogenic glucoside biosynthesis.
Plant J., 111, 2022
|
|
7MNW
 
 | | Crystal Structure of Nup358/RanBP2 Ran-binding domain 1 in complex with Ran-GPPNHP | | Descriptor: | E3 SUMO-protein ligase RanBP2, GTP-binding nuclear protein Ran, MAGNESIUM ION, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNZ
 
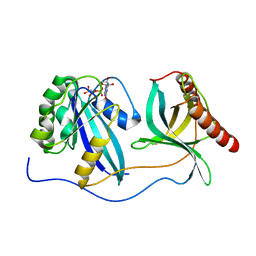 | | Crystal Structure of Nup358/RanBP2 Ran-binding domain 4 in complex with Ran-GPPNHP | | Descriptor: | E3 SUMO-protein ligase RanBP2, GTP-binding nuclear protein Ran, MAGNESIUM ION, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNJ
 
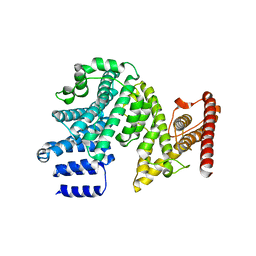 | | Crystal structure of the N-terminal domain of NUP358/RanBP2 (residues 145-673) | | Descriptor: | E3 SUMO-protein ligase RanBP2 | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Patke, A, Dasso, M, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MO0
 
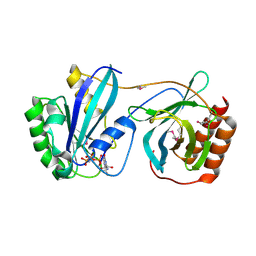 | | Crystal Structure of Nucleoporin NUP50 Ran-Binding Domain in Complex with Ran-GPPNHP | | Descriptor: | GTP-binding nuclear protein Ran, MAGNESIUM ION, Nuclear pore complex protein Nup50, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
5A9G
 
 | | Manganese Superoxide Dismutase from Sphingobacterium sp. T2 | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Rashid, G.M.M, Taylor, C.R, Liu, Y, Zhang, X, Rea, D, Fulop, V, Bugg, T.D.H. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Identification of Manganese Superoxide Dismutase from Sphingobacterium Sp. T2 as a Novel Bacterial Enzyme for Lignin Oxidation.
Acs Chem.Biol., 10, 2015
|
|
3MAT
 
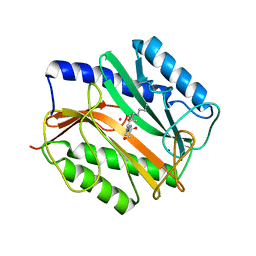 | | E.COLI METHIONINE AMINOPEPTIDASE TRANSITION-STATE INHIBITOR COMPLEX | | Descriptor: | COBALT (II) ION, METHIONINE AMINOPEPTIDASE, SODIUM ION, ... | | Authors: | Lowther, W.T, Orville, A.M, Madden, D.T, Lim, S, Rich, D.H, Matthews, B.W. | | Deposit date: | 1999-03-29 | | Release date: | 1999-06-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Escherichia coli methionine aminopeptidase: implications of crystallographic analyses of the native, mutant, and inhibited enzymes for the mechanism of catalysis.
Biochemistry, 38, 1999
|
|
3M5R
 
 | |
3MIA
 
 | | Crystal structure of HIV-1 Tat complexed with ATP-bound human P-TEFb | | Descriptor: | Cell division protein kinase 9, Cyclin-T1, MAGNESIUM ION, ... | | Authors: | Tahirov, T.H, Babayeva, N.D, Varzavand, K, Cooper, J.J, Sedore, S.C, Price, D.H. | | Deposit date: | 2010-04-09 | | Release date: | 2010-06-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of HIV-1 Tat complexed with human P-TEFb.
Nature, 465, 2010
|
|
5ANY
 
 | | Electron cryo-microscopy of chikungunya virus in complex with neutralizing antibody Fab CHK265 | | Descriptor: | E1, E2, FAB, ... | | Authors: | Fox, J.M, Long, F, Edeling, M.A, Lin, H, Duijl-Richter, M, Fong, R.H, Kahle, K.M, Smit, J.M, Jin, J, Simmons, G, Doranz, B.J, Crowe, J.E, Fremont, D.H, Rossmann, M.G, Diamond, M.S. | | Deposit date: | 2015-09-08 | | Release date: | 2015-11-25 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (16.9 Å) | | Cite: | Broadly Neutralizing Alphavirus Antibodies Bind an Epitope on E2 and Inhibit Entry and Egress.
Cell(Cambridge,Mass.), 163, 2015
|
|
4QDX
 
 | | Crystal structure of Antigen 85C-C209G mutant | | Descriptor: | Diacylglycerol acyltransferase/mycolyltransferase Ag85C, SULFATE ION | | Authors: | Favrot, L, Lajiness, D.H, Ronning, D.R. | | Deposit date: | 2014-05-14 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Inactivation of the Mycobacterium tuberculosis Antigen 85 Complex by Covalent, Allosteric Inhibitors.
J.Biol.Chem., 289, 2014
|
|
4QEK
 
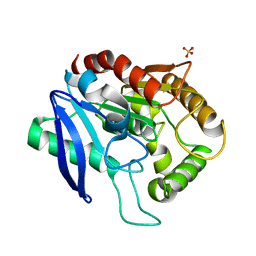 | | Crystal structure of Antigen 85C-S124A mutant | | Descriptor: | Diacylglycerol acyltransferase/mycolyltransferase Ag85C, SULFATE ION, alpha-D-glucopyranose | | Authors: | Favrot, L, Lajiness, D.H, Ronning, D.R. | | Deposit date: | 2014-05-16 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.299 Å) | | Cite: | Inactivation of the Mycobacterium tuberculosis Antigen 85 Complex by Covalent, Allosteric Inhibitors.
J.Biol.Chem., 289, 2014
|
|
4QDZ
 
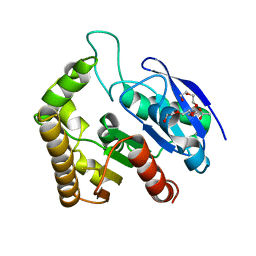 | | Crystal structure of Antigen 85C-E228Q mutant | | Descriptor: | ACETATE ION, Diacylglycerol acyltransferase/mycolyltransferase Ag85C, HEXAETHYLENE GLYCOL | | Authors: | Favrot, L, Lajiness, D.H, Ronning, D.R. | | Deposit date: | 2014-05-14 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.885 Å) | | Cite: | Inactivation of the Mycobacterium tuberculosis Antigen 85 Complex by Covalent, Allosteric Inhibitors.
J.Biol.Chem., 289, 2014
|
|
4QBU
 
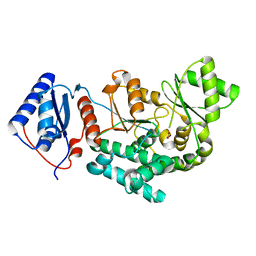 | | Structure of the Acyl Transferase domain of ZmaA | | Descriptor: | FORMIC ACID, ZmaA | | Authors: | Dyer, D.H, Kevany, B.M, Thomas, M.G, Forest, K.T. | | Deposit date: | 2014-05-08 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Polyketide Synthase Acyltransferase Domain Structure Suggests a Recognition Mechanism for Its Hydroxymalonyl-Acyl Carrier Protein Substrate.
Plos One, 9, 2014
|
|
5BWA
 
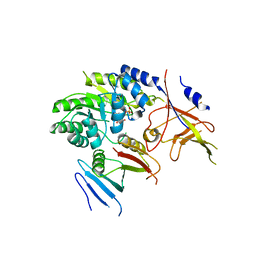 | | Crystal structure of ODC-PLP-AZ1 ternary complex | | Descriptor: | Ornithine decarboxylase, Ornithine decarboxylase antizyme 1, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Wu, D.H. | | Deposit date: | 2015-06-07 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of Ornithine Decarboxylase inactivation and accelerated degradation by polyamine sensor Antizyme1
Sci Rep, 5, 2015
|
|
