6REN
 
 | | Crystal structure of 3fPizza6-SH with Zn2+ | | Descriptor: | 3fPizza6-SH, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
2LIG
 
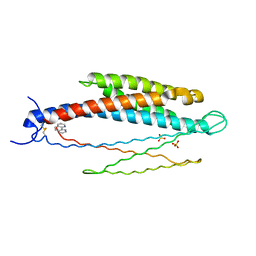 | | THREE-DIMENSIONAL STRUCTURES OF THE LIGAND-BINDING DOMAIN OF THE BACTERIAL ASPARTATE RECEPTOR WITH AND WITHOUT A LIGAND | | Descriptor: | 1,10-PHENANTHROLINE, ASPARTATE RECEPTOR, ASPARTIC ACID, ... | | Authors: | Kim, S.-H, Yeh, J.I, Prive, G.G, Milburn, M, Scott, W, Koshland Junior, D.E. | | Deposit date: | 1995-04-18 | | Release date: | 1995-09-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand.
Science, 254, 1991
|
|
2HG4
 
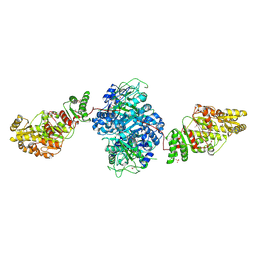 | | Structure of the ketosynthase-acyltransferase didomain of module 5 from DEBS. | | Descriptor: | 6-Deoxyerythronolide B Synthase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Tang, Y, Kim, C.Y, Mathews, I.I, Cane, D.E, Khosla, C. | | Deposit date: | 2006-06-26 | | Release date: | 2006-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The 2.7-A crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2R4B
 
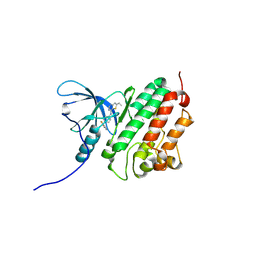 | | ErbB4 kinase domain complexed with a thienopyrimidine inhibitor | | Descriptor: | N-{3-chloro-4-[(3-fluorobenzyl)oxy]phenyl}-6-ethylthieno[3,2-d]pyrimidin-4-amine, Receptor tyrosine-protein kinase erbB-4 | | Authors: | Shewchuk, L.M, Uehling, D.E. | | Deposit date: | 2007-08-31 | | Release date: | 2008-03-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 6-Ethynylthieno[3,2-d]- and 6-ethynylthieno[2,3-d]pyrimidin-4-anilines as tunable covalent modifiers of ErbB kinases.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6THH
 
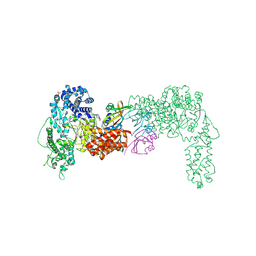 | |
6REH
 
 | | Crystal structure of Pizza6-S with Cu2+ | | Descriptor: | COPPER (II) ION, GLYCEROL, Pizza6-S, ... | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
6REJ
 
 | | Crystal structure of Pizza6-SH with Zn2+ | | Descriptor: | Pizza6-SH, SULFATE ION, ZINC ION | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
6REO
 
 | | Crystal structure of 3fPizza6-SH with Sulphate ion | | Descriptor: | 3fPizza6-SH, SULFATE ION | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Artificial beta-Propeller Protein-based Hydrolases
To Be Published
|
|
6REG
 
 | | Crystal structure of Pizza6-S with Zn2+ | | Descriptor: | GLYCEROL, Pizza6-S, ZINC ION | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
6REK
 
 | | Crystal structure of Pizza6-SH with Cu2+ | | Descriptor: | COPPER (II) ION, GLYCEROL, Pizza6-SH | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
5J7J
 
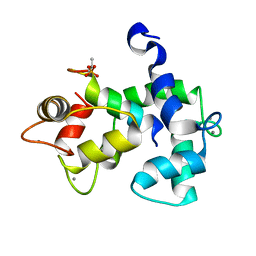 | |
6MG4
 
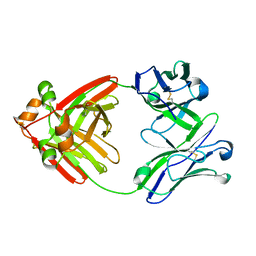 | | Structure of full-length human lambda-6A light chain JTO | | Descriptor: | JTO light chain | | Authors: | Morgan, G.J, Yan, N.L, Mortenson, D.E, Stanfield, R.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2018-09-12 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Stabilization of amyloidogenic immunoglobulin light chains by small molecules.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6MG5
 
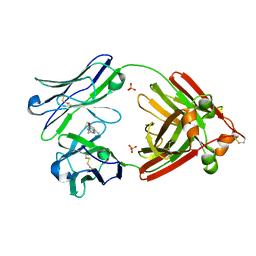 | | Structure of full-length human lambda-6A light chain JTO in complex with coumarin 1 | | Descriptor: | 7-(diethylamino)-4-methyl-2H-1-benzopyran-2-one, Light chain JTO, PHOSPHATE ION | | Authors: | Morgan, G.J, Yan, N.L, Mortenson, D.E, Stanfield, R.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2018-09-12 | | Release date: | 2019-04-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Stabilization of amyloidogenic immunoglobulin light chains by small molecules.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6M9H
 
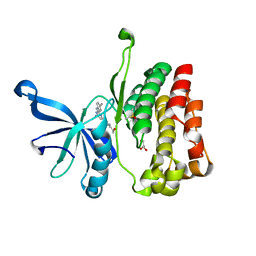 | |
6MG8
 
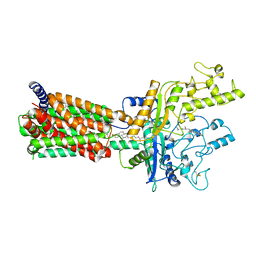 | | Structural basis for cholesterol transport-like activity of the Hedgehog receptor Patched | | Descriptor: | CHOLESTEROL, Protein patched homolog 1 | | Authors: | Zhang, Y, Bulkley, D, Xin, Y, Roberts, K.J, Asarnow, D.E, Sharma, A, Myers, B.R, Cho, W, Cheng, Y, Beachy, P.A. | | Deposit date: | 2018-09-13 | | Release date: | 2018-11-28 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis for Cholesterol Transport-like Activity of the Hedgehog Receptor Patched.
Cell, 175, 2018
|
|
6MPL
 
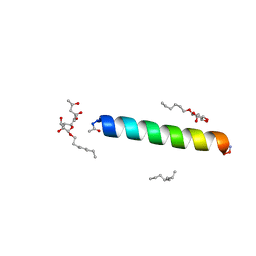 | | Racemic M2-TM I39A crystallized from racemic detergent | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Matrix protein 2, octyl beta-D-glucopyranoside | | Authors: | Kreitler, D.F, Yao, Z, Mortenson, D.E, Forest, K.T, Gellman, S.H. | | Deposit date: | 2018-10-07 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Hendecad Motif Is Preferred for Heterochiral Coiled-Coil Formation.
J. Am. Chem. Soc., 141, 2019
|
|
6MPN
 
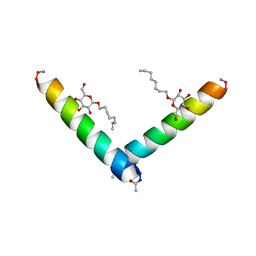 | | Racemic M2-TM I42E crystallized from racemic detergent | | Descriptor: | Matrix protein 2, octyl beta-D-glucopyranoside | | Authors: | Kreitler, D.F, Yao, Z, Mortenson, D.E, Forest, K.T, Gellman, S.H. | | Deposit date: | 2018-10-07 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Hendecad Motif Is Preferred for Heterochiral Coiled-Coil Formation.
J. Am. Chem. Soc., 141, 2019
|
|
6MPM
 
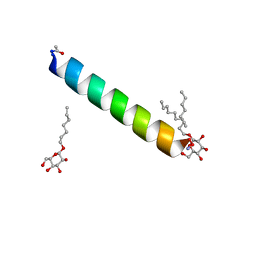 | | Racemic M2-TM I42A crystallized from racemic detergent | | Descriptor: | Matrix protein 2, octyl beta-D-glucopyranoside | | Authors: | Kreitler, D.F, Yao, Z, Mortenson, D.E, Forest, K.T, Gellman, S.H. | | Deposit date: | 2018-10-07 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Hendecad Motif Is Preferred for Heterochiral Coiled-Coil Formation.
J. Am. Chem. Soc., 141, 2019
|
|
6MXT
 
 | | Crystal structure of human beta2 adrenergic receptor bound to salmeterol and Nb71 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Endolysin, ... | | Authors: | Masureel, M, Zou, Y, Picard, L.P, van der Westhuizen, E, Mahoney, J.P, Rodrigues, J.P.G.L.M, Mildorf, T.J, Dror, R.O, Shaw, D.E, Bouvier, M, Pardon, E, Steyaert, J, Sunahara, R.K, Weis, W.I, Zhang, C, Kobilka, B.K. | | Deposit date: | 2018-10-31 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95934224 Å) | | Cite: | Structural insights into binding specificity, efficacy and bias of a beta2AR partial agonist.
Nat. Chem. Biol., 14, 2018
|
|
5K8J
 
 | | Structure of Caulobacter crescentus VapBC1 (apo form) | | Descriptor: | GLYCEROL, Ribonuclease VapC, VapB family protein | | Authors: | Bendtsen, K.L, Xu, K, Luckmann, M, Brodersen, D.E. | | Deposit date: | 2016-05-30 | | Release date: | 2016-12-28 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Toxin inhibition in C. crescentus VapBC1 is mediated by a flexible pseudo-palindromic protein motif and modulated by DNA binding.
Nucleic Acids Res., 45, 2017
|
|
5L1P
 
 | | X-ray Structure of Cytochrome P450 PntM with Pentalenolactone | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Pentalenolactone synthase, pentalenolactone | | Authors: | Duan, L, Jogl, G, Cane, D.E. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | The Cytochrome P450-Catalyzed Oxidative Rearrangement in the Final Step of Pentalenolactone Biosynthesis: Substrate Structure Determines Mechanism.
J.Am.Chem.Soc., 138, 2016
|
|
5L1O
 
 | | X-ray Structure of Cytochrome P450 PntM with Pentalenolactone F | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Pentalenolactone synthase, pentalenolactone F | | Authors: | Duan, L, Jogl, G, Cane, D.E. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Cytochrome P450-Catalyzed Oxidative Rearrangement in the Final Step of Pentalenolactone Biosynthesis: Substrate Structure Determines Mechanism.
J.Am.Chem.Soc., 138, 2016
|
|
5L1Q
 
 | | X-ray Structure of Cytochrome P450 PntM with Dihydropentalenolactone F | | Descriptor: | Dihydropentalenolactone F, PROTOPORPHYRIN IX CONTAINING FE, Pentalenolactone synthase | | Authors: | Duan, L, Jogl, G, Cane, D.E. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Cytochrome P450-Catalyzed Oxidative Rearrangement in the Final Step of Pentalenolactone Biosynthesis: Substrate Structure Determines Mechanism.
J.Am.Chem.Soc., 138, 2016
|
|
5L1S
 
 | |
5L1W
 
 | |
