4NOD
 
 | | Distinct structural features of TFAM drive mitochondrial DNA packaging versus transcriptional activation | | Descriptor: | 5'-D(*CP*CP*AP*AP*CP*CP*AP*AP*GP*CP*CP*CP*CP*AP*TP*AP*CP*CP*CP*CP*AP*A)-3', 5'-D(*TP*TP*GP*GP*GP*GP*TP*AP*TP*GP*GP*GP*GP*CP*TP*TP*GP*GP*(BRU)P*TP*GP*G)-3', Transcription factor A, ... | | Authors: | Ngo, H.B, Lovely, G.A, Phillips, R, Chan, D.C. | | Deposit date: | 2013-11-19 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.897 Å) | | Cite: | Distinct structural features of TFAM drive mitochondrial DNA packaging versus transcriptional activation.
Nat Commun, 5, 2014
|
|
4NPD
 
 | | High-resolution structure of C domain of staphylococcal protein A at cryogenic temperature | | Descriptor: | Immunoglobulin G-binding protein A, THIOCYANATE ION, ZINC ION | | Authors: | Deis, L.N, Pemble IV, C.W, Oas, T.G, Richardson, J.S, Richardson, D.C. | | Deposit date: | 2013-11-21 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Multiscale conformational heterogeneity in staphylococcal protein a: possible determinant of functional plasticity.
Structure, 22, 2014
|
|
4OAH
 
 | |
4OD0
 
 | | Crystal structure of human soluble epoxide hydrolase complexed with 1-(1-propanoylpiperidin-4-yl)-3-[4-(trifluoromethoxy)phenyl]urea | | Descriptor: | 1-(1-propanoylpiperidin-4-yl)-3-[4-(trifluoromethoxy)phenyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Lee, K.S.S, Liu, J, Wagner, K.M, Pakhomova, S, Dong, H, Morisseau, C, Fu, S.H, Yang, J, Wang, P, Ulu, A, Mate, C, Nguyen, L, Wullf, H, Eldin, M.L, Mara, A.A, Newcomer, M.E, Zeldin, D.C, Hammock, B.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Optimized inhibitors of soluble epoxide hydrolase improve in vitro target residence time and in vivo efficacy.
J.Med.Chem., 57, 2014
|
|
4OAF
 
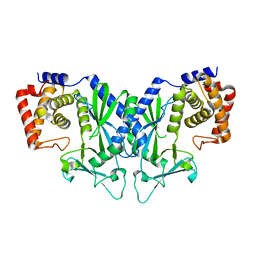 | |
4TQK
 
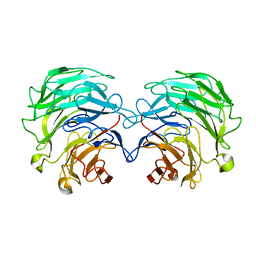 | | Structural basis of specific recognition of non-reducing terminal N-acetylglucosamine by an Agrocybe aegerita lection | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lectin 2 | | Authors: | Hu, Y.L, Ren, X.M, Li, D.F, Jiang, S, Lan, X.Q, Sun, H, Wang, D.C. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Specific Recognition of Non-Reducing Terminal N-Acetylglucosamine by an Agrocybe aegerita Lectin.
Plos One, 10, 2015
|
|
4PR6
 
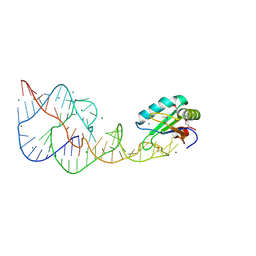 | | A Second Look at the HDV Ribozyme Structure and Dynamics. | | Descriptor: | HDV RIBOZYME SELF-CLEAVED, MAGNESIUM ION, U1 small nuclear ribonucleoprotein A | | Authors: | Kapral, G.J, Jain, S, Noeske, J, Doudna, J.A, Richardson, D.C, Richardson, J.S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-10-29 | | Last modified: | 2014-11-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New tools provide a second look at HDV ribozyme structure, dynamics and cleavage.
Nucleic Acids Res., 42, 2014
|
|
4TQJ
 
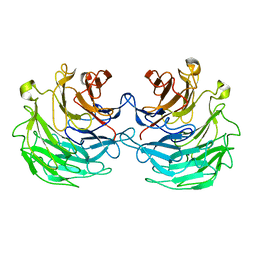 | | Structural basis of specific recognition of non-reducing terminal N-acetylglucosamine by an Agrocybe aegerita lection | | Descriptor: | Lectin 2 | | Authors: | Hu, Y.L, Ren, X.M, Li, D.F, Jiang, S, Lan, X.Q, Sun, H, Wang, D.C. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Specific Recognition of Non-Reducing Terminal N-Acetylglucosamine by an Agrocybe aegerita Lectin.
Plos One, 10, 2015
|
|
4TQM
 
 | | Structural basis of specific recognition of non-reducing terminal N-acetylglucosamine by an Agrocybe aegerita lection | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lectin 2 | | Authors: | Hu, Y.L, Ren, X.M, Li, D.F, Jiang, S, Lan, X.Q, Sun, H, Wang, D.C. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Specific Recognition of Non-Reducing Terminal N-Acetylglucosamine by an Agrocybe aegerita Lectin.
Plos One, 10, 2015
|
|
4MRV
 
 | | Structure of a bacterial Atm1-family ABC transporter | | Descriptor: | ABC transporter related protein, LAURYL DIMETHYLAMINE-N-OXIDE, PHOSPHATE ION, ... | | Authors: | Lee, J.Y, Yang, J.G, Zhitnitsky, D, Lewinson, O, Rees, D.C. | | Deposit date: | 2013-09-17 | | Release date: | 2014-03-19 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for heavy metal detoxification by an Atm1-type ABC exporter.
Science, 343, 2014
|
|
4TKU
 
 | | Reactivated Nitrogenase MoFe-protein from A. vinelandii | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CHLORIDE ION, FE (II) ION, ... | | Authors: | Spatzal, T, Perez, K, Einsle, O, Howard, J.B, Rees, D.C. | | Deposit date: | 2014-05-27 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Ligand binding to the FeMo-cofactor: structures of CO-bound and reactivated nitrogenase.
Science, 345, 2014
|
|
4TKV
 
 | | CO-bound Nitrogenase MoFe-protein from A. vinelandii | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CARBON MONOXIDE, FE (II) ION, ... | | Authors: | Spatzal, T, Perez, K, Einsle, O, Howard, J.B, Rees, D.C. | | Deposit date: | 2014-05-28 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ligand binding to the FeMo-cofactor: structures of CO-bound and reactivated nitrogenase.
Science, 345, 2014
|
|
4P5O
 
 | |
4TN3
 
 | | Structure of the BBox-Coiled-coil region of Rhesus Trim5alpha | | Descriptor: | TRIM5/cyclophilin A fusion protein/T4 Lysozyme chimera, ZINC ION | | Authors: | Kirkpatrick, J.J, Stoye, J.P, Taylor, I.A, Goldstone, D.C. | | Deposit date: | 2014-06-03 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1989 Å) | | Cite: | Structural studies of postentry restriction factors reveal antiparallel dimers that enable avid binding to the HIV-1 capsid lattice.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NPF
 
 | | High-resolution structure of two tandem B domains of staphylococcal protein A connected by the conserved linker | | Descriptor: | Immunoglobulin G-binding protein A | | Authors: | Deis, L.N, Pemble IV, C.W, Oas, T.G, Richardson, J.S, Richardson, D.C. | | Deposit date: | 2013-11-21 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Multiscale conformational heterogeneity in staphylococcal protein a: possible determinant of functional plasticity.
Structure, 22, 2014
|
|
4NA2
 
 | |
4NA1
 
 | |
4NA3
 
 | |
4ND8
 
 | | Av Nitrogenase MoFe Protein High pH Form | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Yang, K.-Y, Haynes, C.A, Spatzal, T, Rees, D.C, Howard, J.B. | | Deposit date: | 2013-10-25 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Turnover-Dependent Inactivation of the Nitrogenase MoFe-Protein at High pH.
Biochemistry, 53, 2014
|
|
4NNU
 
 | | Distinct structural features of TFAM drive mitochondrial DNA packaging versus transcriptional activation | | Descriptor: | DNA1, DNA2, Transcription factor A, ... | | Authors: | Ngo, H.B, Lovely, G.A, Phillips, R, Chan, D.C. | | Deposit date: | 2013-11-19 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Distinct structural features of TFAM drive mitochondrial DNA packaging versus transcriptional activation.
Nat Commun, 5, 2014
|
|
4U3V
 
 | |
4PKX
 
 | | The structure of a conserved Piezo channel domain reveals a novel beta sandwich fold | | Descriptor: | Protein C10C5.1, isoform i | | Authors: | Kamajaya, A, Kaiser, J, Lee, J, Reid, M, Rees, D.C. | | Deposit date: | 2014-05-15 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | The Structure of a Conserved Piezo Channel Domain Reveals a Topologically Distinct beta Sandwich Fold.
Structure, 22, 2014
|
|
4O6P
 
 | | Structural and functional studies the characterization of C58G/C70G mutant in Cys4 Zinc-finger motif in the recombination mediator protein RecR | | Descriptor: | Recombination protein RecR, ZINC ION | | Authors: | Tang, Q, Liu, Y.P, Yan, X.X, Liang, D.C. | | Deposit date: | 2013-12-23 | | Release date: | 2014-12-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional characterization of Cys4 zinc finger motif in the recombination mediator protein RecR.
DNA Repair (Amst.), 24, 2014
|
|
4OU1
 
 | | Crystal structure of a computationally designed retro-aldolase covalently bound to folding probe 1 [(6-methoxynaphthalen-2-yl)(oxiran-2-yl)methanol] | | Descriptor: | (1S,2S)-1-(6-methoxynaphthalen-2-yl)propane-1,2-diol, BENZOIC ACID, PHOSPHATE ION, ... | | Authors: | Bhabha, G, Zhang, X, Ekiert, D.C. | | Deposit date: | 2014-02-14 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Small molecule probes to quantify the functional fraction of a specific protein in a cell with minimal folding equilibrium shifts.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4O6O
 
 | | Structural and functional studies the characterization of Cys4 Zinc-finger motif in the recombination mediator protein RecR | | Descriptor: | IMIDAZOLE, Recombination protein RecR, ZINC ION | | Authors: | Tang, Q, Liu, Y.P, Yan, X.X, Liang, D.C. | | Deposit date: | 2013-12-23 | | Release date: | 2014-12-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional characterization of Cys4 zinc finger motif in the recombination mediator protein RecR.
DNA Repair (Amst.), 24, 2014
|
|
