2GOU
 
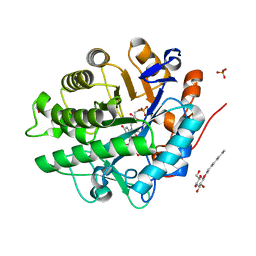 | | Structure of wild type, oxidized SYE1, an OYE homologue from S. oneidensis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Savvides, S.N, van den Hemel, D. | | Deposit date: | 2006-04-14 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Ligand-induced conformational changes in the capping subdomain of a bacterial old yellow enzyme homologue and conserved sequence fingerprints provide new insights into substrate binding.
J.Biol.Chem., 281, 2006
|
|
3U2Z
 
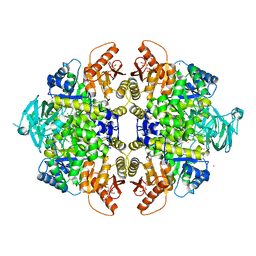 | | Activator-Bound Structure of Human Pyruvate Kinase M2 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-(3-aminobenzyl)-4-methyl-2-methylsulfinyl-4,6-dihydro-5H-thieno[2',3':4,5]pyrrolo[2,3-d]pyridazin-5-one, Pyruvate kinase isozymes M1/M2, ... | | Authors: | Hong, B, Dimov, S, Tempel, W, Auld, D, Thomas, C, Boxer, M, Jianq, J.-K, Skoumbourdis, A, Min, S, Southall, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Inglese, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-10-04 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis.
Nat.Chem.Biol., 8, 2012
|
|
3GWL
 
 | |
3V88
 
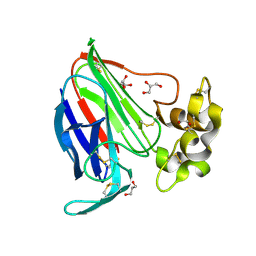 | | Thaumatin by Classical Hanging Drop Vapour Diffusion after 18.1 MGy X-Ray dose at ESRF ID29 beamline (Best Case) | | Descriptor: | GLYCEROL, Thaumatin I | | Authors: | Belmonte, L, Scudieri, D, Tripathi, S, Pechkova, E, Nicolini, C. | | Deposit date: | 2011-12-22 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Langmuir-Blodgett nanotemplate and radiation resistance in protein crystals: state of the art.
CRIT.REV.EUKARYOT.GENE EXPR., 22, 2012
|
|
2GQA
 
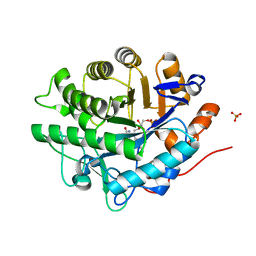 | | Structure of NADH-reduced SYE1, an OYE homologue from S. oneidensis | | Descriptor: | FLAVIN MONONUCLEOTIDE, SULFATE ION, oxidoreductase, ... | | Authors: | Savvides, S.N, van den Hemel, D. | | Deposit date: | 2006-04-20 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ligand-induced conformational changes in the capping subdomain of a bacterial old yellow enzyme homologue and conserved sequence fingerprints provide new insights into substrate binding.
J.Biol.Chem., 281, 2006
|
|
5T4O
 
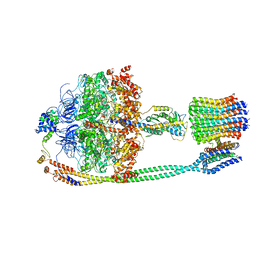 | | Autoinhibited E. coli ATP synthase state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Sobti, M, Smits, C, Wong, A.S.W, Ishmukhametov, R, Stock, D, Sandin, S, Stewart, A.G. | | Deposit date: | 2016-08-29 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Cryo-EM structures of the autoinhibitedE. coliATP synthase in three rotational states.
Elife, 5, 2016
|
|
2GUZ
 
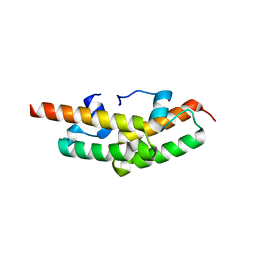 | | Structure of the Tim14-Tim16 complex of the mitochondrial protein import motor | | Descriptor: | CITRATE ANION, Mitochondrial import inner membrane translocase subunit TIM14, Mitochondrial import inner membrane translocase subunit TIM16 | | Authors: | Mokranjac, D, Bourenkov, G, Hell, K, Neupert, W, Groll, M. | | Deposit date: | 2006-05-02 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of Tim14 and Tim16, the J and J-like components of the mitochondrial protein import motor.
Embo J., 25, 2006
|
|
3VBU
 
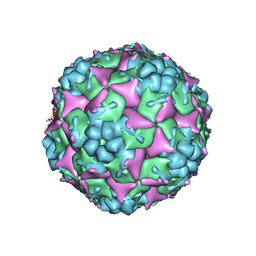 | | Crystal structure of empty human Enterovirus 71 particle | | Descriptor: | Genome Polyprotein, capsid protein VP0, capsid protein VP1, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5T74
 
 | | Human carboanhydrase F131C_C206S double mutant in complex with 14 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[2,5-bis(oxidanylidene)pyrrol-1-yl]-~{N}-(4-sulfamoylphenyl)ethanamide, 4-(HYDROXYMERCURY)BENZOIC ACID, ... | | Authors: | DuBay, K.H, Iwan, K, Osorio-Planes, L, Geissler, P, Groll, M, Trauner, D, Broichhagen, J. | | Deposit date: | 2016-09-02 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A Predictive Approach for the Optical Control of Carbonic Anhydrase II Activity.
ACS Chem. Biol., 13, 2018
|
|
5W6M
 
 | |
3FGB
 
 | | Crystal structure of the Q89ZH8_BACTN protein from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium target BtR289b. | | Descriptor: | uncharacterized protein Q89ZH8_BACTN | | Authors: | Vorobiev, S.M, Lew, S, Seetharaman, J, Lee, D, Foote, L.E, Ciccosanti, C, Janjua, H, Xiao, R, Acton, T, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-05 | | Release date: | 2008-12-23 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the Q89ZH8_BACTN protein from Bacteroides thetaiotaomicron.
To be Published
|
|
3FI8
 
 | | Crystal structure of choline kinase from Plasmodium Falciparum, PF14_0020 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Choline kinase, MAGNESIUM ION, ... | | Authors: | Wernimont, A.K, Pizarro, J.C, Artz, J.D, Amaya, M.F, Xiao, T, Lew, J, Wasney, G, Senesterra, G, Kozieradzki, I, Cossar, D, Vedadi, M, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-11 | | Release date: | 2009-02-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of choline kinase from Plasmodium Falciparum, PF14_0020
TO BE PUBLISHED
|
|
2H16
 
 | | Structure of human ADP-ribosylation factor-like 5 (ARL5) | | Descriptor: | ADP-ribosylation factor-like protein 5A, GUANOSINE-5'-DIPHOSPHATE, UNKNOWN ATOM OR ION | | Authors: | Rabeh, W.M, Tempel, W, Yaniw, D, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-16 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human ADP-ribosylation factor-like 5 (ARL5)
To be Published
|
|
2GZO
 
 | | NMR structure of UPF0301 PROTEIN SO3346 from Shewanella oneidensis: Northeast Structural Genomics Consortium target SOR39 | | Descriptor: | UPF0301 protein SO3346 | | Authors: | Singarapu, K.K, Liu, G, Eletsky, A, Xu, D, Sukumaran, D.K, Mei, J, Xiao, R, Cunningham, K, Ma, L.C, Ritu, S, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-05-11 | | Release date: | 2006-06-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of UPF0301 PROTEIN SO3346 from Shewanella oneidensis: Northeast Structural Genomics Consortium target SOR39
To be Published
|
|
3VN4
 
 | | Crystal structure of the exosite-containing fragment of human ADAMTS13 (P475S mutant) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A disintegrin and metalloproteinase with thrombospondin motifs 13, ... | | Authors: | Nakayama, D, Akiyama, M, Takeda, S, Kokame, K, Takagi, J, Miyata, T. | | Deposit date: | 2011-12-21 | | Release date: | 2012-12-26 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure and enzymatic activity of an ADAMTS-13 mutant with the East Asian-specific P475S polymorphism.
J.Thromb.Haemost., 11, 2013
|
|
3V7M
 
 | | Crystal structure of monoclonal human anti-Rhesus D Fc IgG1 T125(YB2/0) in the presence of Zn2+ | | Descriptor: | GLYCEROL, IMIDAZOLE, Ig gamma-1 chain C region, ... | | Authors: | Menez, R, Stura, E.A, Bourel, D, Siberil, S, Jorieux, S, De Romeuf, C, Ducancel, F, Fridman, W.H, Teillaud, J.L. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Effect of zinc on human IgG1 and its Fc gamma R interactions.
Immunol.Lett., 143, 2012
|
|
3R37
 
 | | Crystal structure of Arthrobacter sp. strain SU 4-hydroxybenzoyl CoA thioesterase mutant E73Q complexed with 4-hydroxyphenacyl CoA | | Descriptor: | 4-HYDROXYPHENACYL COENZYME A, 4-hydroxybenzoyl-CoA thioesterase | | Authors: | Holden, H.M, Thoden, J.B, Song, F, Zhuang, Z, Trujillo, M, Dunaway-Mariano, D. | | Deposit date: | 2011-03-15 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Catalytic Mechanism of the Hotdog-fold Enzyme Superfamily 4-Hydroxybenzoyl-CoA Thioesterase from Arthrobacter sp. Strain SU.
Biochemistry, 51, 2012
|
|
3V8B
 
 | | Crystal Structure of a 3-ketoacyl-ACP reductase from Sinorhizobium meliloti 1021 | | Descriptor: | Putative dehydrogenase, possibly 3-oxoacyl-[acyl-carrier protein] reductase | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-22 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of a 3-ketoacyl-ACP reductase from Sinorhizobium meliloti 1021
To be Published
|
|
2H85
 
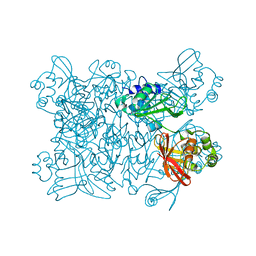 | | Crystal Structure of Nsp 15 from SARS | | Descriptor: | Putative orf1ab polyprotein | | Authors: | Ricagno, S, Egloff, M.P, Ulferts, R, Coutard, B, Nurizzo, D, Campanacci, V, Cambillau, C, Ziebuhr, J, Canard, B. | | Deposit date: | 2006-06-06 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5W3W
 
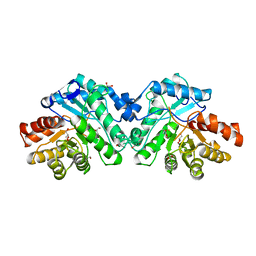 | | Crystal structure of SsoPox AsD6 mutant (V27A-Y97W-L228M-W263M) - open form | | Descriptor: | 1,2-ETHANEDIOL, Aryldialkylphosphatase, COBALT (II) ION, ... | | Authors: | Hiblot, J, Gotthard, G, Jacquet, P, Daude, D, Bergonzi, C, Chabriere, E, Elias, M. | | Deposit date: | 2017-06-08 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Rational engineering of a native hyperthermostable lactonase into a broad spectrum phosphotriesterase.
Sci Rep, 7, 2017
|
|
3VCA
 
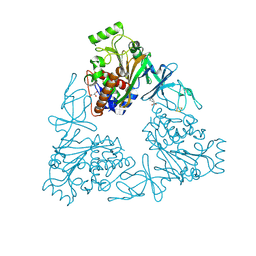 | | Quaternary Ammonium Oxidative Demethylation: X-ray Crystallographic, Resonance Raman and UV-visible Spectroscopic Analysis of a Rieske-type Demethylase | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, ... | | Authors: | Daughtry, K.D, Xiao, Y, Stoner-Ma, D, Cho, E, Orville, A.M, Liu, P, Allen, K.N. | | Deposit date: | 2012-01-03 | | Release date: | 2012-02-08 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Quaternary Ammonium Oxidative Demethylation: X-ray Crystallographic, Resonance Raman, and UV-Visible Spectroscopic Analysis of a Rieske-Type Demethylase.
J.Am.Chem.Soc., 134, 2012
|
|
2HBH
 
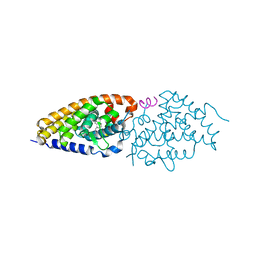 | | Crystal structure of Vitamin D nuclear receptor ligand binding domain bound to a locked side-chain analog of calcitriol and SRC-1 peptide | | Descriptor: | 1,3-CYCLOHEXANEDIOL, 4-METHYLENE-5-[(2E)-[(1S,3AS,7AS)-OCTAHYDRO-1-(5-HYDROXY-5-METHYL-1,3-HEXADIYNYL)-7A-METHYL-4H-INDEN-4-YLIDENE]ETHYLIDENE]-, (1R,3S,5Z), ... | | Authors: | Rochel, N, Hourai, S, Moras, D. | | Deposit date: | 2006-06-14 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the vitamin D nuclear receptor ligand binding domain in complex with a locked side chain analog of calcitriol
Arch.Biochem.Biophys., 460, 2007
|
|
3VNV
 
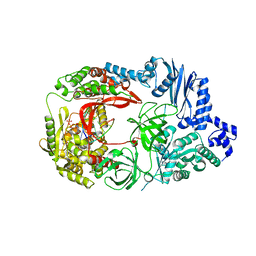 | | Complex structure of viral RNA polymerase II | | Descriptor: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, Elongation factor Ts, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2012-01-18 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Mechanism for template-independent terminal adenylation activity of Q beta replicase
Structure, 20, 2012
|
|
2HC4
 
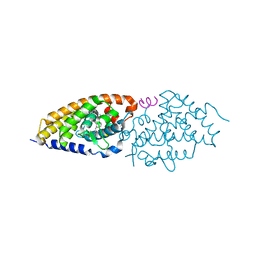 | | Crystal structure of the LBD of VDR of Danio rerio in complex with calcitriol | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, SRC-1 from Nuclear receptor coactivator 1, Vitamin D receptor | | Authors: | Ciesielski, F, Rochel, N, Moras, D. | | Deposit date: | 2006-06-15 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Adaptability of the Vitamin D nuclear receptor to the synthetic ligand Gemini: remodelling the LBP with one side chain rotation
J.Steroid Biochem.Mol.Biol., 103, 2007
|
|
2HCD
 
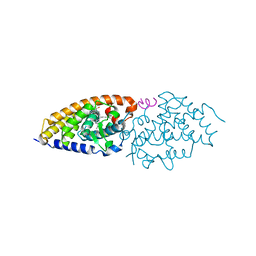 | | Crystal structure of the ligand binding domain of the Vitamin D nuclear receptor in complex with Gemini and a coactivator peptide | | Descriptor: | 21-NOR-9,10-SECOCHOLESTA-5,7,10(19)-TRIENE-1,3,25-TRIOL, 20-(4-HYDROXY-4-METHYLPENTYL)-, (1A,3B,5Z,7E), ... | | Authors: | Ciesielski, F, Rochel, N, Moras, D. | | Deposit date: | 2006-06-16 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Adaptability of the Vitamin D nuclear receptor to the synthetic ligand Gemini: remodelling the LBP with one side chain rotation
J.Steroid Biochem.Mol.Biol., 103, 2007
|
|
