8ASG
 
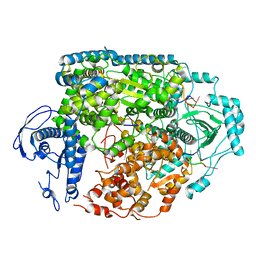 | | Structure of the SFTSV L protein bound in a resting state [RESTING] | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*AP*CP*AP*CP*AP*GP*AP*GP*AP*CP*GP*CP*CP*CP*AP*GP*AP*UP*GP*A*)-3'), RNA (5'-R(*GP*AP*UP*CP*UP*GP*GP*GP*CP*GP*GP*UP*CP*UP*UP*UP*GP*UP*GP*U*)-3'), ... | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Milewski, M, Busch, C, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2022-08-19 | | Release date: | 2023-01-18 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into viral genome replication by the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 51, 2023
|
|
2UWP
 
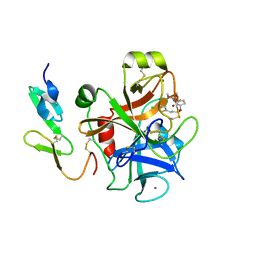 | | Factor Xa inhibitor complex | | Descriptor: | 2-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}ETHANESULFONAMIDE, CALCIUM ION, COAGULATION FACTOR X, ... | | Authors: | Young, R.J, Brown, D, Burns-Kurtis, C.L, Chan, C, Convery, M.A, Hubbard, J.A, Kelly, H.A, Pateman, A.J, Patikis, A, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Zhou, P, Thorpe, J.H. | | Deposit date: | 2007-03-22 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Selective and Dual Action Orally Active Inhibitors of Thrombin and Factor Xa.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
8ASB
 
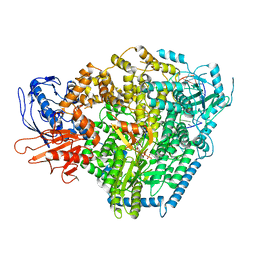 | | Structure of the SFTSV L protein stalled at early elongation with the endonuclease domain in a raised conformation [EARLY-ELONGATION-ENDO] | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, MAGNESIUM ION, RNA (5'-R(*AP*CP*AP*CP*AP*GP*AP*GP*AP*CP*GP*CP*CP*CP*AP*GP*A)-3'), ... | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Milewski, M, Busch, C, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2022-08-18 | | Release date: | 2023-01-18 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into viral genome replication by the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 51, 2023
|
|
8ASD
 
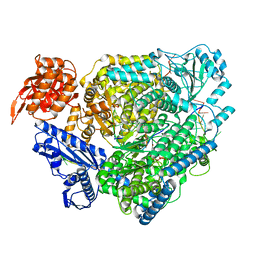 | | Structure of the SFTSV L protein stalled at late elongation [LATE-ELONGATION] | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, MAGNESIUM ION, RNA (5'-R(P*AP*AP*AP*AP*AP*AP*GP*AP*UP*CP*UP*GP*GP*GP*CP*GP*GP*UP*CP*UP*UP*UP*GP*UP*GP*U)-3'), ... | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Milewski, M, Busch, C, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2022-08-19 | | Release date: | 2023-01-18 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into viral genome replication by the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 51, 2023
|
|
8AS6
 
 | | Structure of the SFTSV L protein bound to 5' cRNA hook [5' HOOK] | | Descriptor: | RNA (5'-R(*AP*CP*AP*CP*AP*GP*AP*GP*AP*CP*GP*CP*CP*C)-3'), RNA-dependent RNA-polymerase L protein | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Milewski, M, Busch, C, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2022-08-18 | | Release date: | 2023-01-18 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into viral genome replication by the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 51, 2023
|
|
8AS7
 
 | | Structure of the SFTSV L protein stalled at early elongation [EARLY-ELONGATION] | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, MAGNESIUM ION, ... | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Milewski, M, Busch, C, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2022-08-18 | | Release date: | 2023-01-18 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into viral genome replication by the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 51, 2023
|
|
8AJX
 
 | | E. coli NfsA with Fumarate | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, FUMARIC ACID, ... | | Authors: | Day, M.A, Jarrom, D, White, S.A, Hyde, E.I. | | Deposit date: | 2022-07-28 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Oxygen-insensitive nitroreductase E. coli NfsA, but not NfsB, is inhibited by fumarate.
Proteins, 91, 2023
|
|
7NR6
 
 | | Structure of NUDT15 in complex with NSC56456 | | Descriptor: | 1,2-ETHANEDIOL, 2-azanyl-9-cyclohexyl-3~{H}-purine-6-thione, MAGNESIUM ION, ... | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2021-03-03 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Coupling cellular target engagement to drug-induced responses with CeTEAM
To Be Published
|
|
8AI7
 
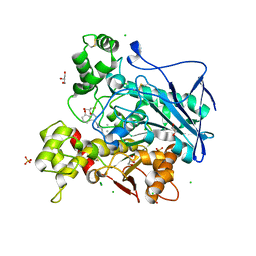 | | Structure of carbamoylated human butyrylcholinesterase upon reaction with 3-(((2-cycloheptylethyl)(methyl)amino)methyl)-1H-indol-7-yl N,N-dimethylcarbamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[[2-cycloheptylethyl(methyl)amino]methyl]-1~{H}-indol-7-ol, ... | | Authors: | Brazzolotto, X, Meden, A, Knez, D, Gobec, S, Nachon, F. | | Deposit date: | 2022-07-25 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Pseudo-irreversible butyrylcholinesterase inhibitors: Structure-activity relationships, computational and crystallographic study of the N-dialkyl O-arylcarbamate warhead.
Eur.J.Med.Chem., 247, 2023
|
|
2V9N
 
 | |
2UZY
 
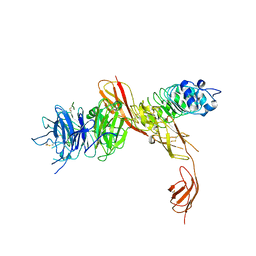 | | Structure of the human receptor tyrosine kinase Met in complex with the Listeria monocytogenes invasion protein inlb: low resolution, Crystal form II | | Descriptor: | HEPATOCYTE GROWTH FACTOR RECEPTOR, INTERNALIN B | | Authors: | Niemann, H.H, Jager, V, Butler, P.J.G, van den Heuvel, J, Schmidt, S, Ferraris, D, Gherardi, E, Heinz, D.W. | | Deposit date: | 2007-05-02 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of the Human Receptor Tyrosine Kinase met in Complex with the Listeria Invasion Protein Inlb
Cell(Cambridge,Mass.), 130, 2007
|
|
8AJ7
 
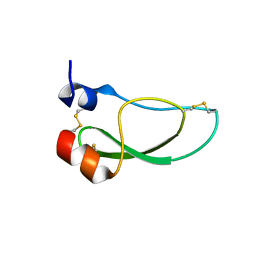 | | Kunitz domain of Amblyomin-X | | Descriptor: | 1,2-ETHANEDIOL, Kunitz domain of Amblyomin-X | | Authors: | Ciccone, L, Servent, D, Stura, E.A. | | Deposit date: | 2022-07-27 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional properties of the Kunitz-type and C-terminal domains of Amblyomin-X supporting its antitumor activity.
Front Mol Biosci, 10, 2023
|
|
2V3W
 
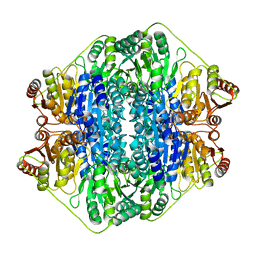 | | Crystal structure of the benzoylformate decarboxylase variant L461A from Pseudomonas putida | | Descriptor: | BENZOYLFORMATE DECARBOXYLASE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Gocke, D, Walter, L, Gauchenova, K, Kolter, G, Knoll, M, Berthold, C.L, Schneider, G, Pleiss, J, Mueller, M, Pohl, M. | | Deposit date: | 2007-06-25 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Protein Design of Thdp-Dependent Enzymes-Engineering Stereoselectivity.
Chembiochem, 9, 2008
|
|
1PJ5
 
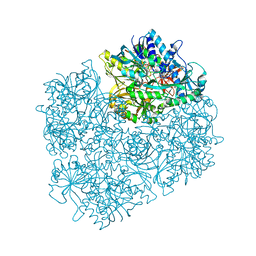 | | Crystal structure of dimethylglycine oxidase of Arthrobacter globiformis in complex with acetate | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, N,N-dimethylglycine oxidase, ... | | Authors: | Leys, D, Basran, J, Scrutton, N.S. | | Deposit date: | 2003-06-01 | | Release date: | 2003-10-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Channelling and formation of 'active' formaldehyde in dimethylglycine oxidase.
Embo J., 22, 2003
|
|
1PG7
 
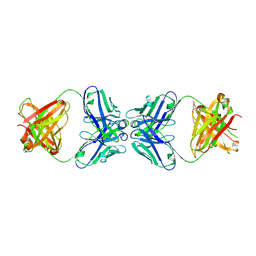 | | Murine 6A6 Fab in complex with humanized anti-Tissue Factor D3H44 Fab | | Descriptor: | humanized antibody D3H44, murine antibody 6A6 Fab fragment | | Authors: | Eigenbrot, C, Meng, Y.G, Krishnamurthy, R, Lipari, M.T, Presta, L, Devaux, B, Wong, T, Moran, P, Bullens, S, Kirchhofer, D. | | Deposit date: | 2003-05-27 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into how an anti-idiotypic antibody against D3H44 (anti-tissue factor antibody) restores normal coagulation.
J.Mol.Biol., 331, 2003
|
|
2UUO
 
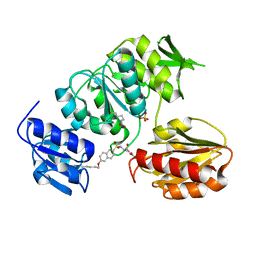 | | Crystal structure of MurD ligase in complex with D-Glu containing sulfonamide inhibitor | | Descriptor: | N-{[6-(PENTYLOXY)NAPHTHALEN-2-YL]SULFONYL}-D-GLUTAMIC ACID, SULFATE ION, UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE | | Authors: | Humljan, J, Kotnik, M, Contreras-Martel, C, Blanot, D, Urleb, U, Dessen, A, Solmajer, T, Gobec, S. | | Deposit date: | 2007-03-06 | | Release date: | 2008-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel naphthalene-N-sulfonyl-D-glutamic acid derivatives as inhibitors of MurD, a key peptidoglycan biosynthesis enzyme.
J. Med. Chem., 51, 2008
|
|
2UWE
 
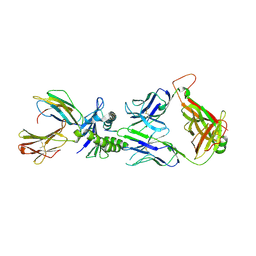 | | Large CDR3a loop alteration as a function of MHC mutation | | Descriptor: | AHIII TCR ALPHA CHAIN, AHIII TCR BETA CHAIN, BETA-2-MICROGLOBULIN, ... | | Authors: | Miller, P.J, Pazy, Y, Conti, B, Riddle, D, Biddison, W.E, Appella, E, Collins, E.J. | | Deposit date: | 2007-03-20 | | Release date: | 2007-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Single Mhc Mutation Eliminates Enthalpy Associated with T Cell Receptor Binding.
J.Mol.Biol., 373, 2007
|
|
7O7M
 
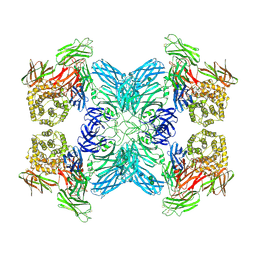 | | (h-alpha2M)4 native II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-2-macroglobulin, ... | | Authors: | Luque, D, Goulas, T, Mata, C.P, Mendes, S.R, Gomis-Ruth, F.X, Caston, J.R. | | Deposit date: | 2021-04-13 | | Release date: | 2022-04-13 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cryo-EM structures show the mechanistic basis of pan-peptidase inhibition by human alpha 2 -macroglobulin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1PAV
 
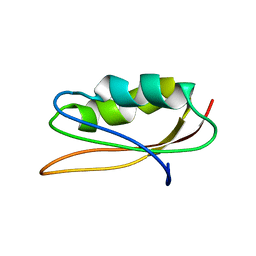 | | SOLUTION NMR STRUCTURE OF HYPOTHETICAL PROTEIN TA1414 OF THERMOPLASMA ACIDOPHILUM | | Descriptor: | Hypothetical protein Ta1170/Ta1414 | | Authors: | Monleon, D, Yee, A, Liu, C.S, Arrowsmith, C, Celda, B. | | Deposit date: | 2003-05-14 | | Release date: | 2003-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hypothetical protein TA1414 from Thermoplasma acidophilum.
J.Biomol.Nmr, 28, 2004
|
|
7O2L
 
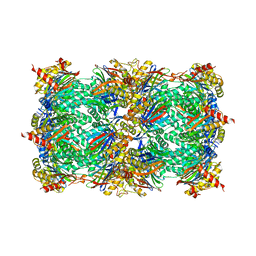 | | Yeast 20S proteasome in complex with the covalently bound inhibitor b-lactone (2R,3S)-3-isopropyl-4-oxo-2-oxetane-carboxylate (IOC) | | Descriptor: | (2 {R},3 {S})-3-methanoyl-4-methyl-2-hydroxy-pentanoic acid, 20S proteasome, BJ4_G0020160.mRNA.1.CDS.1, ... | | Authors: | Shi, Y.M, Hirschmann, M, Shi, Y.N, Shabbir, A, Abebew, D, Tobias, N.J, Gruen, P, Crames, J.J, Poeschel, L, Kuttenlochner, W, Richter, C, Herrmann, J, Mueller, R, Thanwisai, A, Pidot, S.J, Stinear, T.P, Groll, M, Kim, Y, Bode, H. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Global analysis of biosynthetic gene clusters reveals conserved and unique natural products in entomopathogenic nematode-symbiotic bacteria.
Nat.Chem., 14, 2022
|
|
1PBH
 
 | |
7O3D
 
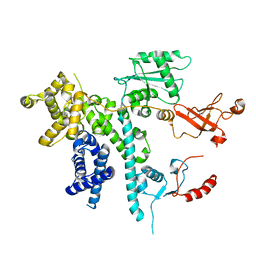 | | Cooperation between the intrinsically disordered and ordered regions of Spt6 regulates nucleosome and Pol II CTD binding, and nucleosome assembly | | Descriptor: | Transcription elongation factor SPT6 | | Authors: | Kasiliauskaite, A, Kubicek, K, Klumpler, T, Zanova, M, Zapletal, D, Novacek, J, Stefl, R. | | Deposit date: | 2021-04-01 | | Release date: | 2022-04-13 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Cooperation between intrinsically disordered and ordered regions of Spt6 regulates nucleosome and Pol II CTD binding, and nucleosome assembly.
Nucleic Acids Res., 50, 2022
|
|
7O7R
 
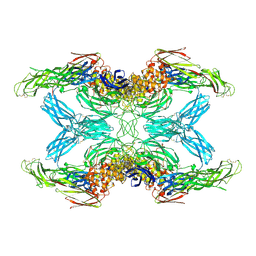 | | (h-alpha2M)4 plasmin-activated I state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-2-macroglobulin, ... | | Authors: | Luque, D, Goulas, T, Mata, C.P, Mendes, S.R, Gomis-Ruth, F.X, Caston, J.R. | | Deposit date: | 2021-04-13 | | Release date: | 2022-04-13 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures show the mechanistic basis of pan-peptidase inhibition by human alpha 2 -macroglobulin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8B0F
 
 | | CryoEM structure of C5b8-CD59 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Bubeck, D, Couves, E.C, Gardner, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for membrane attack complex inhibition by CD59.
Nat Commun, 14, 2023
|
|
8B0G
 
 | | 2C9, C5b9-CD59 structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD59 glycoprotein, Complement C5, ... | | Authors: | Couves, E.C, Gardner, S, Bubeck, D. | | Deposit date: | 2022-09-07 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for membrane attack complex inhibition by CD59.
Nat Commun, 14, 2023
|
|
