1K5O
 
 | | CPI-17(35-120) deletion mutant | | Descriptor: | CPI-17 | | Authors: | Ohki, S, Eto, M, Kariya, E, Hayano, T, Hayashi, Y, Yazawa, M, Brautigan, D, Kainosho, M. | | Deposit date: | 2001-10-11 | | Release date: | 2002-10-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Myosin Phosphatase Inhibitor Protein CPI-17 Shows Phosphorylation-induced Conformational Changes Responsible for Activation
J.Mol.Biol., 314, 2001
|
|
7Q6I
 
 | | Vibrio maritimus FtsA 1-396 ATP and FtsN 1-29, bent tetramers in double filament arrangement | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division protein FtsA, Cell division protein FtsN (polyAla model), ... | | Authors: | Nierhaus, T, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2021-11-07 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Bacterial divisome protein FtsA forms curved antiparallel double filaments when binding to FtsN.
Nat Microbiol, 7, 2022
|
|
5JKI
 
 | | Crystal structure of the first transmembrane PAP2 type phosphatidylglycerolphosphate phosphatase from Bacillus subtilis | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Putative lipid phosphate phosphatase YodM, TUNGSTATE(VI)ION, ... | | Authors: | El Ghachi, M, Howe, N, Lampion, A, Delbrassine, F, Vogeley, L, Caffrey, M, Sauvage, E, Auger, R, Guiseppe, A, Roure, S, Perlier, S, Mengin-lecreulx, D, Foglino, M, Touze, T. | | Deposit date: | 2016-04-26 | | Release date: | 2017-02-22 | | Last modified: | 2017-05-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure and biochemical characterization of the transmembrane PAP2 type phosphatidylglycerol phosphate phosphatase from Bacillus subtilis.
Cell. Mol. Life Sci., 74, 2017
|
|
1K5W
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE SYNAPTOTAGMIN 1 C2B-DOMAIN: SYNAPTOTAGMIN 1 AS A PHOSPHOLIPID BINDING MACHINE | | Descriptor: | CALCIUM ION, Synaptotagmin I | | Authors: | Fernandez, I, Arac, D, Ubach, J, Gerber, S.H, Shin, O, Gao, Y, Anderson, R.G.W, Sudhof, T.C, Rizo, J. | | Deposit date: | 2001-10-12 | | Release date: | 2002-01-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the synaptotagmin 1 C2B-domain: synaptotagmin 1 as a phospholipid binding machine.
Neuron, 32, 2001
|
|
1YJ1
 
 | | X-ray Crystal Structure of a Chemically Synthesized [D-Gln35]Ubiquitin | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ubiquitin | | Authors: | Bang, D, Makhatadze, G.I, Tereshko, V, Kossiakoff, A.A, Kent, S.B. | | Deposit date: | 2005-01-13 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | X-ray Crystal Structure of a Chemically Synthesized [D-Gln35]Ubiquitin
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
7Q6G
 
 | | Xenorhabdus poinarii FtsA 1-396 ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division protein FtsA, MAGNESIUM ION | | Authors: | Nierhaus, T, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2021-11-07 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bacterial divisome protein FtsA forms curved antiparallel double filaments when binding to FtsN.
Nat Microbiol, 7, 2022
|
|
7Q71
 
 | | The crystallographic structure of the Ligand Binding domain of the NR7 nuclear receptor from the amphioxus Branchiostoma lanceolatum | | Descriptor: | CHLORIDE ION, Nuclear hormone receptor 7, PHOSPHATE ION | | Authors: | Billas, I.M.L, McEwen, A.G, Hazemann, I, Moras, D, Laudet, V. | | Deposit date: | 2021-11-09 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel nuclear receptor subfamily enlightens the origin of heterodimerization.
Bmc Biol., 20, 2022
|
|
7Q6F
 
 | | Vibrio maritimus FtsA 1-396 ATP, double filament | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division protein FtsA, MAGNESIUM ION | | Authors: | Nierhaus, T, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2021-11-07 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Bacterial divisome protein FtsA forms curved antiparallel double filaments when binding to FtsN.
Nat Microbiol, 7, 2022
|
|
1I5H
 
 | |
1YJE
 
 | | Crystal structure of the rNGFI-B ligand-binding domain | | Descriptor: | Orphan nuclear receptor NR4A1 | | Authors: | Flaig, R, Greschik, H, Peluso-Iltis, C, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-14 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the cell-specific activities of the NGFI-B and the Nurr1 ligand-binding domain.
J.Biol.Chem., 280, 2005
|
|
4HCY
 
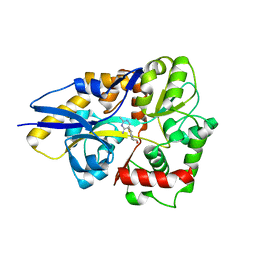 | | Structure of a eukaryotic thiaminase-I bound to the thiamin analogue 3-deazathiamin | | Descriptor: | 2-{4-[(4-amino-2-methylpyrimidin-5-yl)methyl]-3-methylthiophen-2-yl}ethanol, thiaminase-I | | Authors: | Kreinbring, C.A, Hubbard, P.A, Leeper, F.J, Hawksley, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2012-10-01 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of a eukaryotic thiaminase I.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5IZS
 
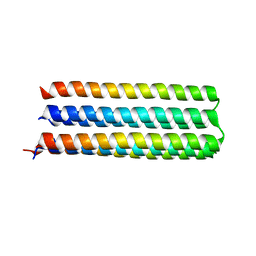 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | Descriptor: | Designed protein 5L6HC3_1 | | Authors: | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Oberdorfer, G. | | Deposit date: | 2016-03-25 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
5JKG
 
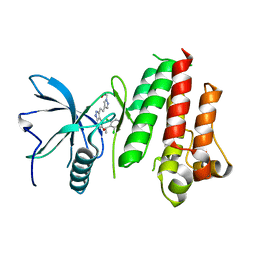 | | The crystal structure of FGFR4 kinase domain in complex with LY2874455 | | Descriptor: | 2-[4-[E-2-[5-[(1R)-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-1H-indazol-3-yl]ethenyl]pyrazol-1-yl]ethanol, Fibroblast growth factor receptor 4 | | Authors: | Wu, D, Chen, L, Chen, Y. | | Deposit date: | 2016-04-26 | | Release date: | 2016-10-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | Crystal Structure of the FGFR4/LY2874455 Complex Reveals Insights into the Pan-FGFR Selectivity of LY2874455
Plos One, 11, 2016
|
|
6JF1
 
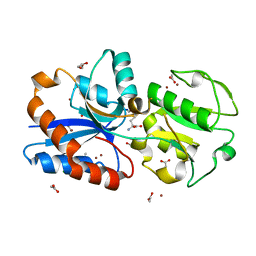 | | Crystal structure of the substrate binding protein of a methionine transporter from Streptococcus pneumoniae | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jha, B, Vyas, R, Bhushan, J, Sehgal, D, Biswal, B.K. | | Deposit date: | 2019-02-07 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into the substrate specificity of SP_0149, the substrate-binding protein of a methionine ABC transporter from Streptococcus pneumoniae.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
5J0K
 
 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | Descriptor: | designed protein 2L4HC2_23 | | Authors: | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Groves, B, Langan, R.A, Oberdorfer, G, Ford, A, Gilmore, J, Xu, C, DiMaio, F, Seelig, G. | | Deposit date: | 2016-03-28 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
5IU4
 
 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with ZM241385 at 1.7A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Segala, E, Guo, D, Cheng, R.K.Y, Bortolato, A, Deflorian, F, Dore, A.S, Errey, J.C, Heitman, L.H, Ijzerman, A.P, Marshall, F.H, Cooke, R.M. | | Deposit date: | 2016-03-17 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Controlling the Dissociation of Ligands from the Adenosine A2A Receptor through Modulation of Salt Bridge Strength.
J.Med.Chem., 59, 2016
|
|
1YJO
 
 | | Structure of NNQQNY from yeast prion Sup35 with zinc acetate | | Descriptor: | ACETIC ACID, Eukaryotic peptide chain release factor GTP-binding subunit, ZINC ION | | Authors: | Nelson, R, Sawaya, M.R, Balbirnie, M, Madsen, A.O, Riekel, C, Grothe, R, Eisenberg, D. | | Deposit date: | 2005-01-15 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of the cross-beta spine of amyloid-like fibrils.
Nature, 435, 2005
|
|
1YJP
 
 | | Structure of GNNQQNY from yeast prion Sup35 | | Descriptor: | Eukaryotic peptide chain release factor GTP-binding subunit | | Authors: | Nelson, R, Sawaya, M.R, Balbirnie, M, Madsen, A.O, Riekel, C, Grothe, R, Eisenberg, D. | | Deposit date: | 2005-01-15 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the cross-beta spine of amyloid-like fibrils.
Nature, 435, 2005
|
|
6JHR
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F6 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-18 | | Release date: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
6SXW
 
 | | Crystal structure of the first RRM domain of human Zinc finger protein 638 (ZNF638) | | Descriptor: | SULFATE ION, Zinc finger protein 638 | | Authors: | Newman, J.A, Aitkenhead, H, Wang, D, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-09-26 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Crystal structure of the first RRM domain of human Zinc finger protein 638 (ZNF638)
To Be Published
|
|
6JMF
 
 | |
1W8W
 
 | | CBM29-2 mutant Y46A: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | NON-CATALYTIC PROTEIN 1 | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-09-30 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
6IWG
 
 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*05104 complexed with N-myristoylated 4-mer lipopeptide derived from SIV nef protein | | Descriptor: | 1,2-ETHANEDIOL, BORIC ACID, Beta-2-microglobulin, ... | | Authors: | Yamamoto, Y, Morita, D, Sugita, M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and Structure of an MHC Class I-Encoded Protein with the Potential to PresentN-Myristoylated 4-mer Peptides to T Cells.
J Immunol., 202, 2019
|
|
1ICF
 
 | | CRYSTAL STRUCTURE OF MHC CLASS II ASSOCIATED P41 II FRAGMENT IN COMPLEX WITH CATHEPSIN L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (CATHEPSIN L: HEAVY CHAIN), PROTEIN (CATHEPSIN L: LIGHT CHAIN), ... | | Authors: | Guncar, G, Pungercic, G, Klemencic, I, Turk, V, Turk, D. | | Deposit date: | 1999-01-07 | | Release date: | 2000-01-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of MHC class II-associated p41 Ii fragment bound to cathepsin L reveals the structural basis for differentiation between cathepsins L and S.
EMBO J., 18, 1999
|
|
6IZR
 
 | | Whole structure of a 15-stranded ParM filament from Clostridium botulinum | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative plasmid segregation protein ParM | | Authors: | Koh, F, Narita, A, Lee, L.J, Tan, Y.Z, Dandey, V.P, Tanaka, K, Popp, D, Robinson, R.C. | | Deposit date: | 2018-12-20 | | Release date: | 2019-06-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | The structure of a 15-stranded actin-like filament from Clostridium botulinum.
Nat Commun, 10, 2019
|
|
