4DDW
 
 | |
4DJZ
 
 | | Catalytic fragment of masp-1 in complex with its specific inhibitor developed by directed evolution on sgci scaffold | | Descriptor: | Mannan-binding lectin serine protease 1 heavy chain, Mannan-binding lectin serine protease 1 light chain, Protease inhibitor SGPI-2 | | Authors: | Heja, D, Harmat, V, Fodor, K, Wilmanns, M, Dobo, J, Kekesi, K.A, Zavodszky, P, Gal, P, Pal, G. | | Deposit date: | 2012-02-03 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Monospecific Inhibitors Show That Both Mannan-binding Lectin-associated Serine Protease-1 (MASP-1) and -2 Are Essential for Lectin Pathway Activation and Reveal Structural Plasticity of MASP-2.
J.Biol.Chem., 287, 2012
|
|
4DLO
 
 | | Crystal structure of the GAIN and HormR domains of brain angiogenesis inhibitor 3 (BAI3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Brain-specific angiogenesis inhibitor 3, ... | | Authors: | Arac, D, Boucard, A.A, Bolliger, M.F, Nguyen, J, Soltis, M, Sudhof, T.C, Brunger, A.T. | | Deposit date: | 2012-02-06 | | Release date: | 2012-02-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis.
Embo J., 31, 2012
|
|
4DNV
 
 | | Crystal structure of the W285F mutant of UVB-resistance protein UVR8 | | Descriptor: | AT5g63860/MGI19_6 | | Authors: | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Zhang, J, Wang, J, Shi, Y. | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
4DTH
 
 | | Structure of a VgrG Vibrio cholerae toxin ACD domain in complex with ATP and Mg++ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Durand, E, Audoly, G, Derrez, E, Spinelli, S, Ortiz-Lombardia, M, Cascales, E, Raoult, D, Cambillau, C. | | Deposit date: | 2012-02-21 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of a VgrG Vibrio cholerae toxin ACD domain in complex with ATP and Mg++
J.Biol.Chem., 2012
|
|
4DP3
 
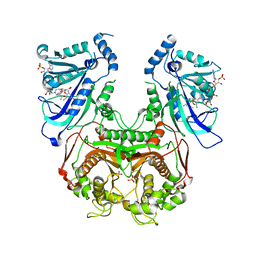 | | Quadruple mutant (N51I+C59R+S108N+I164L) plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with P218 and NADPH | | Descriptor: | 3-(2-{3-[(2,4-diamino-6-ethylpyrimidin-5-yl)oxy]propoxy}phenyl)propanoic acid, Bifunctional dihydrofolate reductase-thymidylate synthase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Yuthavong, Y, Vilaivan, T, Kamchonwongpaisan, S, Charman, S.A, McLennan, D.N, White, K.L, Vivas, L, Bongard, E, Chitnumsub, P, Tarnchompoo, B, Thongphanchang, C, Taweechai, S, Vanichtanakul, J, Arwon, U, Fantauzzi, P, Yuvaniyama, J, Charman, W.N, Matthews, D. | | Deposit date: | 2012-02-13 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DSY
 
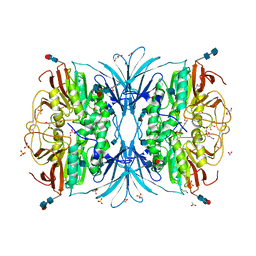 | | Crystal structure of red kidney bean purple acid phosphatase in complex with Maybridge fragment CC24201 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-phenylpyridine-3-carboxylic acid, ... | | Authors: | Feder, D, Hussein, W.M, Clayton, D.J, Kan, M, Schenk, G, McGeary, R.P, Guddat, L.W. | | Deposit date: | 2012-02-20 | | Release date: | 2012-09-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of purple acid phosphatase inhibitors by fragment-based screening: promising new leads for osteoporosis therapeutics.
Chem.Biol.Drug Des., 80, 2012
|
|
4DCC
 
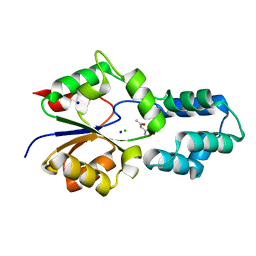 | | Crystal structure of had family enzyme bt-2542 from bacteroides thetaiotaomicron (target efi-501088) | | Descriptor: | CHLORIDE ION, Putative haloacid dehalogenase-like hydrolase, SODIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-17 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of protein Bt-2542 from Bacteroides Thetaiotaomicron (Target Efi-501088)
To be Published
|
|
4DCL
 
 | | Computationally Designed Self-assembling tetrahedron protein, T308, Crystallized in space group F23 | | Descriptor: | Putative acetyltransferase SACOL2570 | | Authors: | Sawaya, M.R, King, N.P, Sheffler, W, Baker, D, Yeates, T.O. | | Deposit date: | 2012-01-17 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Computational design of self-assembling protein nanomaterials with atomic level accuracy.
Science, 336, 2012
|
|
4DCU
 
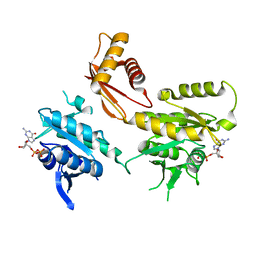 | | Crystal Structure of B. subtilis EngA in complex with GDP | | Descriptor: | GTP-BINDING PROTEIN ENGA, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Reiser, J.-B, Housset, D, Foucher, A.-E, Jault, J.-M. | | Deposit date: | 2012-01-18 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potassium Acts as a GTPase-Activating Element on Each Nucleotide-Binding Domain of the Essential Bacillus subtilis EngA.
Plos One, 7, 2012
|
|
4DAM
 
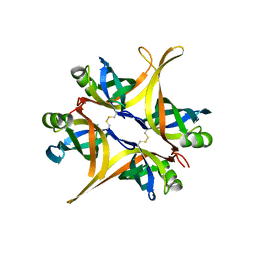 | | Crystal structure of small single-stranded DNA-binding protein from Streptomyces coelicolor | | Descriptor: | Single-stranded DNA-binding protein 1 | | Authors: | Filic, Z, Herron, P, Ivic, N, Luic, M, Manjasetty, B.A, Paradzik, T, Vujaklija, D. | | Deposit date: | 2012-01-13 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-function relationships of two paralogous single-stranded DNA-binding proteins from Streptomyces coelicolor: implication of SsbB in chromosome segregation during sporulation.
Nucleic Acids Res., 41, 2013
|
|
4DG5
 
 | |
4E1D
 
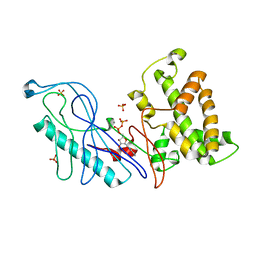 | | Structure of a VgrG Vibrio cholerae toxin ACD domain in complex with ADP and Mn++ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Durand, E, Audoly, G, Derrez, E, Spinelli, S, Ortiz-Lombardia, M, Cascales, E, Raoult, D, Cambillau, C. | | Deposit date: | 2012-03-06 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure and functional characterization of the Vibrio cholerae toxin
from the VgrG/MARTX family.
J.Biol.Chem., 2012
|
|
4DD0
 
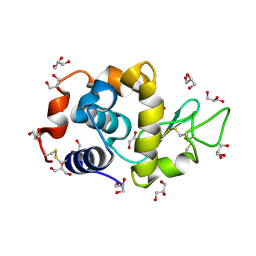 | | EVAL processed HEWL, cisplatin aqueous glycerol | | Descriptor: | GLYCEROL, Lysozyme C | | Authors: | Tanley, S.W, Schreurs, A.M, Kroon-Batenburg, L.M, Meredith, J, Prendergast, R, Walsh, D, Bryant, P, Levy, C, Helliwell, J.R. | | Deposit date: | 2012-01-18 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of the effect that dimethyl sulfoxide (DMSO) has on cisplatin and carboplatin binding to histidine in a protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DJI
 
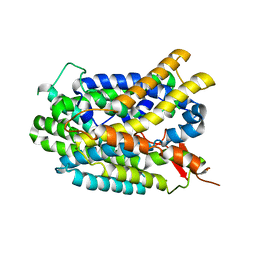 | | Structure of glutamate-GABA antiporter GadC | | Descriptor: | Probable glutamate/gamma-aminobutyrate antiporter | | Authors: | Ma, D, Lu, P.L, Yan, C.Y, Fan, C, Yin, P, Wang, J.W, Shi, Y.G. | | Deposit date: | 2012-02-02 | | Release date: | 2012-03-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.187 Å) | | Cite: | Structure and mechanism of a glutamate-GABA antiporter
Nature, 483, 2012
|
|
4DLQ
 
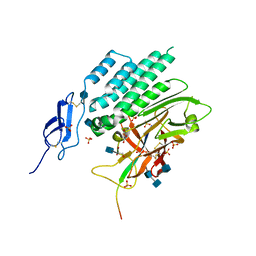 | | Crystal structure of the GAIN and HormR domains of CIRL 1/Latrophilin 1 (CL1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Arac, D, Boucard, A.A, Bolliger, M.F, Nguyen, J, Soltis, M, Sudhof, T.C, Brunger, A.T. | | Deposit date: | 2012-02-06 | | Release date: | 2012-02-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis.
Embo J., 31, 2012
|
|
4DLI
 
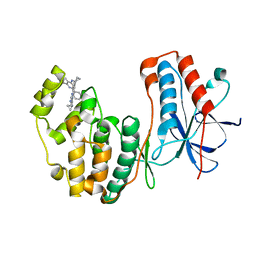 | | Human p38 MAP kinase in complex with RL87 | | Descriptor: | Mitogen-activated protein kinase 14, N~4~-cyclopropyl-2-phenylquinazoline-4,7-diamine | | Authors: | Gruetter, C, Getlik, M, Simard, J.R, Rauh, D. | | Deposit date: | 2012-02-06 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Fluorophore labeled kinase detects ligands that bind within the MAPK insert of p38alpha kinase.
Plos One, 7, 2012
|
|
4DQ9
 
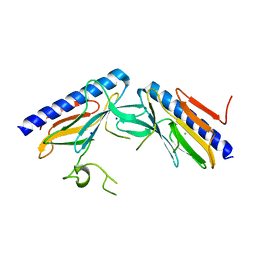 | | Crystal structure of the minor pseudopilin EPSH from the type II secretion system of Vibrio cholerae | | Descriptor: | CHLORIDE ION, General secretion pathway protein H, SODIUM ION | | Authors: | Raghunathan, K, Vago, F.S, Grindem, D, Ball, T, Wedemeyer, W.J, Arvidson, D.N. | | Deposit date: | 2012-02-15 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The 1.59 angstrom resolution structure of the minor pseudopilin EpsH of Vibrio cholerae reveals a long flexible loop.
Biochim.Biophys.Acta, 1844, 2013
|
|
4DM9
 
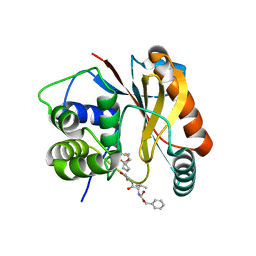 | | The Crystal Structure of Ubiquitin Carboxy-terminal hydrolase L1 (UCHL1) bound to a tripeptide fluoromethyl ketone Z-VAE(OMe)-FMK | | Descriptor: | Tripeptide fluoromethyl ketone inhibitor Z-VAE(OMe)-FMK, Ubiquitin carboxyl-terminal hydrolase isozyme L1 | | Authors: | Davies, C.W, Chaney, J, Korbel, G, Ringe, D, Petsko, G.A, Ploegh, H, Das, C. | | Deposit date: | 2012-02-07 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The co-crystal structure of ubiquitin carboxy-terminal hydrolase L1 (UCHL1) with a tripeptide fluoromethyl ketone (Z-VAE(OMe)-FMK).
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6Y1C
 
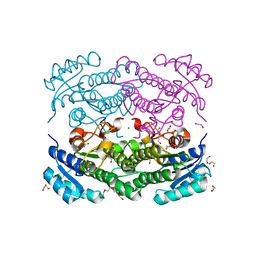 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant D54F | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hermann, J, Bischoff, D, Janowski, R, Niessing, D, Grob, P, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2020-02-11 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Controlling Protein Crystallization by Free Energy Guided Design of Interactions at Crystal Contacts
Crystals, 11, 2021
|
|
2JQW
 
 | |
4M35
 
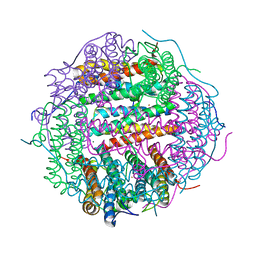 | | Crystal structure of gated-pore mutant H126/141D of second DNA-Binding protein under starvation from Mycobacterium smegmatis | | Descriptor: | CHLORIDE ION, FE (II) ION, MAGNESIUM ION, ... | | Authors: | Williams, S.M, Chandran, A.V, Vijayabaskar, M.S, Roy, S, Balaram, H, Vishveshwara, S, Vijayan, M, Chatterji, D. | | Deposit date: | 2013-08-06 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A histidine aspartate ionic lock gates the iron passage in miniferritins from Mycobacterium smegmatis
J.Biol.Chem., 289, 2014
|
|
2JQ0
 
 | | Phylloseptin-1 | | Descriptor: | Phylloseptin-1 | | Authors: | Resende, J.M, Mendonca Moraes, C, Almeida, F.C.L, Prates, M.V, Cesar, A, Valente, A, Bemquerer, M.P, Pilo-Veloso, D, Bechinger, B. | | Deposit date: | 2007-05-25 | | Release date: | 2008-06-03 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structures of the antimicrobial peptides phylloseptin-1, -2, and -3 and biological activity: The role of charges and hydrogen bonding interactions in stabilizing helix conformations
Peptides, 29, 2008
|
|
2JQN
 
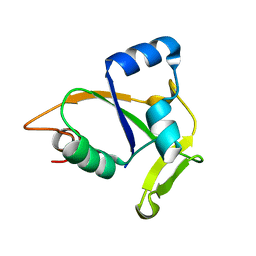 | | Solution NMR structure of CC0527 from Caulobacter crescentus. Northeast Structural Genomics Target CCR55 | | Descriptor: | Hypothetical protein | | Authors: | Aramini, J.M, Rossi, P, Moseley, H.N.B, Wang, D, Nwosu, C, Cunningham, K, Ma, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-05 | | Release date: | 2007-07-03 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of CC0527 from Caulobacter crescentus
To be Published
|
|
2JOM
 
 | |
