1WTF
 
 | | Crystal structure of Bacillus thermoproteolyticus Ferredoxin Variants Containing Unexpected [3Fe-4S] Cluster that is linked to Coenzyme A at 1.6 A Resolution | | Descriptor: | COENZYME A, FE3-S4 CLUSTER, Ferredoxin, ... | | Authors: | Shirakawa, T, Takahashi, Y, Wada, K, Hirota, J, Takao, T, Ohmori, D, Fukuyama, K. | | Deposit date: | 2004-11-22 | | Release date: | 2005-11-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of variant molecules of Bacillus thermoproteolyticus ferredoxin: crystal structure reveals bound coenzyme A and an unexpected [3Fe-4S] cluster associated with a canonical [4Fe-4S] ligand motif
Biochemistry, 44, 2005
|
|
1WWE
 
 | | NMR Structure Determined for MLV NC complex with RNA Sequence UUUUGCU | | Descriptor: | 5'-R(P*UP*UP*UP*UP*GP*CP*U)-3', Nucleoprotein p10, ZINC ION | | Authors: | Dey, A, York, D, Smalls-Mantey, A, Summers, M.F. | | Deposit date: | 2005-01-05 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Composition and sequence-dependent binding of RNA to the nucleocapsid protein of Moloney murine leukemia virus(,)
Biochemistry, 44, 2005
|
|
3VT0
 
 | | Crystal structure of Ct1,3Gal43A in complex with lactose | | Descriptor: | GLYCEROL, Ricin B lectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
5OQV
 
 | | Near-atomic resolution fibril structure of complete amyloid-beta(1-42) by cryo-EM | | Descriptor: | Amyloid beta A4 protein | | Authors: | Gremer, L, Schoelzel, D, Schenk, C, Reinartz, E, Labahn, J, Ravelli, R, Tusche, M, Lopez-Iglesias, C, Hoyer, W, Heise, H, Willbold, D, Schroeder, G.F. | | Deposit date: | 2017-08-14 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Fibril structure of amyloid-beta (1-42) by cryo-electron microscopy.
Science, 358, 2017
|
|
4E0L
 
 | | FYLLYYT segment from human Beta 2 Microglobulin (62-68) displayed on 54-membered macrocycle scaffold | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cyclic pseudo-peptide FYLLYYT(ORN)KN(HAO)SA(ORN) | | Authors: | Zhao, M, Liu, C, Michael, S.R, Eisenberg, D. | | Deposit date: | 2012-03-04 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Out-of-register beta-sheets suggest a pathway to toxic amyloid aggregates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1WBR
 
 | |
3VW7
 
 | | Crystal structure of human protease-activated receptor 1 (PAR1) bound with antagonist vorapaxar at 2.2 angstrom | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Proteinase-activated receptor 1, ... | | Authors: | Zhang, C, Srinivasan, Y, Arlow, D.H, Fung, J.J, Palmer, D, Zheng, Y, Green, H.F, Pandey, A, Dror, R.O, Shaw, D.E, Weis, W.I, Coughlin, S.R, Kobilka, B.K. | | Deposit date: | 2012-08-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-resolution crystal structure of human protease-activated receptor 1
Nature, 492, 2012
|
|
1WCU
 
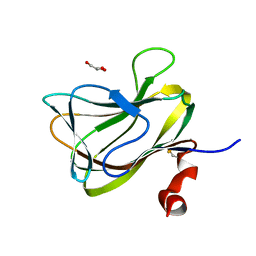 | | CBM29_1, A Family 29 Carbohydrate Binding Module from Piromyces equi | | Descriptor: | GLYCEROL, NON-CATALYTIC PROTEIN 1 | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davis, G.J, Gilbert, H.J. | | Deposit date: | 2004-11-22 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1X99
 
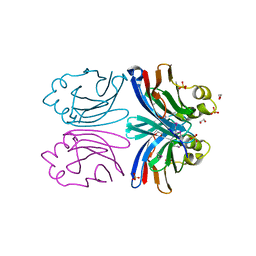 | | X-ray crystal structure of Xerocomus chrysenteron lectin XCL at 1.4 Angstroms resolution, mutated at Q46M, V54M, L58M | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, lectin | | Authors: | Birck, C, Damian, L, Marty-Detraves, C, Lougarre, A, Schulze-Briese, C, Koehl, P, Fournier, D, Paquereau, L, Samama, J.P. | | Deposit date: | 2004-08-20 | | Release date: | 2004-12-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A New Lectin Family with Structure Similarity to Actinoporins Revealed by the Crystal Structure of Xerocomus chrysenteron Lectin XCL
J.Mol.Biol., 344, 2004
|
|
3VSZ
 
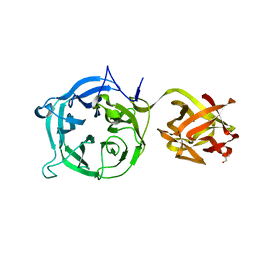 | | Crystal structure of Ct1,3Gal43A in complex with galactan | | Descriptor: | GLYCEROL, Ricin B lectin, beta-D-galactopyranose-(1-3)-beta-D-galactopyranose, ... | | Authors: | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.893 Å) | | Cite: | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
2MBX
 
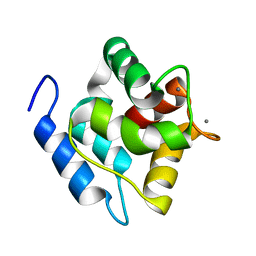 | | Structure, dynamics and stability of allergen cod parvalbumin Gad m 1 by solution and high-pressure NMR. | | Descriptor: | CALCIUM ION, Parvalbumin beta | | Authors: | Moraes, A.H, Ackerbauer, D, Bublin, M, Ferreira, F, Almeida, F.C.L, Breiteneder, H, Valente, A. | | Deposit date: | 2013-08-07 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution and high-pressure NMR studies of the structure, dynamics, and stability of the cross-reactive allergenic cod parvalbumin Gad m 1.
Proteins, 82, 2014
|
|
4MB9
 
 | | Structure of Streptococcus pneumonia ParE in complex with AZ13102335 | | Descriptor: | 1-ethyl-3-{6-(pyrimidin-5-yl)-5-[(3R)-tetrahydrofuran-3-ylmethoxy][1,3]thiazolo[5,4-b]pyridin-2-yl}urea, DNA topoisomerase IV, B subunit, ... | | Authors: | Ogg, D, Tucker, J. | | Deposit date: | 2013-08-19 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Thiazolopyridine Ureas as Novel Antitubercular Agents Acting through Inhibition of DNA Gyrase B.
J.Med.Chem., 56, 2013
|
|
3VSF
 
 | | Crystal structure of 1,3Gal43A, an exo-beta-1,3-Galactanase from Clostridium thermocellum | | Descriptor: | GLYCEROL, Ricin B lectin | | Authors: | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | Deposit date: | 2012-04-25 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.757 Å) | | Cite: | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
4MBC
 
 | | Structure of Streptococcus pneumonia ParE in complex with AZ13053807 | | Descriptor: | 1-{5-[2-(morpholin-4-yl)ethoxy]-6-(pyridin-3-yl)[1,3]thiazolo[5,4-b]pyridin-2-yl}-3-prop-2-en-1-ylurea, DNA topoisomerase IV, B subunit | | Authors: | Ogg, D, Tucker, J. | | Deposit date: | 2013-08-19 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Thiazolopyridine Ureas as Novel Antitubercular Agents Acting through Inhibition of DNA Gyrase B.
J.Med.Chem., 56, 2013
|
|
1XBU
 
 | | Streptomyces griseus aminopeptidase complexed with p-iodo-D-phenylalanine | | Descriptor: | Aminopeptidase, CALCIUM ION, P-IODO-D-PHENYLALANINE, ... | | Authors: | Reiland, V, Gilboa, R, Spungin-Bialik, A, Schomburg, D, Shoham, Y, Blumberg, S, Shoham, G. | | Deposit date: | 2004-08-31 | | Release date: | 2005-10-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Streptomyces griseus aminopeptidase complexed with p-iodo-D-phenylalanine
To be Published
|
|
1XA4
 
 | | Crystal structure of CaiB, a type III CoA transferase in carnitine metabolism | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COENZYME A, Crotonobetainyl-CoA:carnitine CoA-transferase, ... | | Authors: | Stenmark, P, Gurmu, D, Nordlund, P. | | Deposit date: | 2004-08-25 | | Release date: | 2004-11-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of CaiB, a Type-III CoA Transferase in Carnitine Metabolism
Biochemistry, 43, 2004
|
|
1XDV
 
 | | Experimentally Phased Structure of Human the Son of Sevenless protein at 4.1 Ang. | | Descriptor: | Son of sevenless protein homolog 1 | | Authors: | Sondermann, H, Soisson, S.M, Boykevisch, S, Yang, S.S, Bar-Sagi, D, Kuriyan, J. | | Deposit date: | 2004-09-08 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural analysis of autoinhibition in the ras activator son of sevenless.
Cell(Cambridge,Mass.), 119, 2004
|
|
1X8D
 
 | | Crystal structure of E. coli YiiL protein containing L-rhamnose | | Descriptor: | Hypothetical protein yiiL, L-RHAMNOSE | | Authors: | Ryu, K.S, Kim, J.I, Cho, S.J, Park, D, Park, C, Lee, J.O, Choi, B.S. | | Deposit date: | 2004-08-18 | | Release date: | 2005-05-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into the Monosaccharide Specificity of Escherichia coli Rhamnose Mutarotase
J.Mol.Biol., 349, 2005
|
|
1XB8
 
 | | Zn substituted form of D62C/K74C double mutant of Pseudomonas Aeruginosa Azurin | | Descriptor: | Azurin, ZINC ION | | Authors: | Tigerstrom, A, Schwarz, F, Karlsson, G, Okvist, M, Alvarez-Rua, C, Maeder, D, Robb, F.T, Sjolin, L. | | Deposit date: | 2004-08-30 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of a novel disulfide bond and engineered electrostatic interactions on the thermostability of azurin
Biochemistry, 43, 2004
|
|
6SDG
 
 | |
1X70
 
 | | HUMAN DIPEPTIDYL PEPTIDASE IV IN COMPLEX WITH A BETA AMINO ACID INHIBITOR | | Descriptor: | (2R)-4-OXO-4-[3-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[4,3-A]PYRAZIN-7(8H)-YL]-1-(2,4,5-TRIFLUOROPHENYL)BUTAN-2-A MINE, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kim, D, Wang, L, Beconi, M, Eiermann, G.J, Fisher, M.H, He, H, Hickey, G.J, Leiting, B, Lyons, K. | | Deposit date: | 2004-08-12 | | Release date: | 2005-01-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | (2R)-4-Oxo-4-[3-(Trifluoromethyl)-5,6-dihydro[1,2,4]triazolo[4,3-a]pyrazin- 7(8H)-yl]-1-(2,4,5-trifluorophenyl)butan-2-amine: A Potent, Orally Active Dipeptidyl Peptidase IV Inhibitor for the Treatment of Type 2 Diabetes
J.Med.Chem., 48, 2005
|
|
3VVW
 
 | | NDP52 in complex with LC3C | | Descriptor: | Calcium-binding and coiled-coil domain-containing protein 2, Microtubule-associated proteins 1A/1B light chain 3C | | Authors: | Akutsu, M, Muhlinen, N.V, Randow, F, Komander, D. | | Deposit date: | 2012-07-28 | | Release date: | 2013-02-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy
Mol.Cell, 48, 2012
|
|
7DWW
 
 | | Crystal structure of the computationally designed msDPBB_sym2 protein | | Descriptor: | msDPBB_sym2 protein | | Authors: | Yagi, S, Schiex, T, Vucinic, J, Barbe, S, Simoncini, D, Tagami, S. | | Deposit date: | 2021-01-18 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Seven Amino Acid Types Suffice to Create the Core Fold of RNA Polymerase.
J.Am.Chem.Soc., 143, 2021
|
|
1XB6
 
 | | The K24R mutant of Pseudomonas Aeruginosa Azurin | | Descriptor: | Azurin, COPPER (II) ION | | Authors: | Tigerstrom, A, Schwarz, F, Karlsson, G, Okvist, M, Alvarez-Rua, C, Maeder, D, Robb, F.T, Sjolin, L. | | Deposit date: | 2004-08-30 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.823 Å) | | Cite: | Effects of a novel disulfide bond and engineered electrostatic interactions on the thermostability of azurin
Biochemistry, 43, 2004
|
|
1XCV
 
 | |
