2B4F
 
 | | Structure Of A Cold-Adapted Family 8 Xylanase in complex with substrate | | Descriptor: | beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, endo-1,4-beta-xylanase | | Authors: | De Vos, D, Collins, T, Savvides, S.N, Feller, G, Van Beeumen, J.J. | | Deposit date: | 2005-09-23 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Oligosaccharide binding in family 8 glycosidases: crystal structures of active-site mutants of the beta-1,4-xylanase pXyl from Pseudoaltermonas haloplanktis TAH3a in complex with substrate and product.
Biochemistry, 45, 2006
|
|
2AZM
 
 | | Crystal structure of the MDC1 brct repeat in complex with the histone tail of gamma-H2AX | | Descriptor: | GAMMA-H2AX HISTONE, Mediator of DNA damage checkpoint protein 1 | | Authors: | Clapperton, J.A, Stucki, M, Mohammad, D, Yaffe, M.B, Jackson, S.P, Smerdon, S.J. | | Deposit date: | 2005-09-12 | | Release date: | 2006-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | MDC1 Directly Binds Phosphorylated Histone H2AX to Regulate Cellular Responses to DNA Double-Strand Breaks
Cell(Cambridge,Mass.), 123, 2005
|
|
2AH1
 
 | | Crystal structure of aromatic amine dehydrogenase (AADH) from Alcaligenes faecalis | | Descriptor: | Aromatic amine dehydrogenase | | Authors: | Masgrau, L, Roujeinikova, A, Johannissen, L.O, Hothi, P, Basran, J, Ranaghan, K.E, Mulholland, A.J, Sutcliffe, M.J, Scrutton, N.S, Leys, D. | | Deposit date: | 2005-07-27 | | Release date: | 2006-04-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomic description of an enzyme reaction dominated by proton tunneling
Science, 312, 2006
|
|
2AII
 
 | | wild-type Formylglycine generating enzyme reacted with iodoacetamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETAMIDE, CALCIUM ION, ... | | Authors: | Roeser, D, Rudolph, M.G. | | Deposit date: | 2005-07-29 | | Release date: | 2005-12-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | A general binding mechanism for all human sulfatases by the formylglycine-generating enzyme
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2AJM
 
 | | NMR structure of the in-plane membrane anchor domain [1-28] of the monotopic NonStructural Protein 5A (NS5A) from the Bovine Viral Diarrhea Virus (BVDV) | | Descriptor: | Nonstructural protein 5A | | Authors: | Sapay, N, Montserret, R, Chipot, C, Brass, V, Moradpour, D, Deleage, G, Penin, F. | | Deposit date: | 2005-08-02 | | Release date: | 2005-08-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure and molecular dynamics of the in-plane membrane anchor of nonstructural protein 5A from bovine viral diarrhea virus.
Biochemistry, 45, 2006
|
|
2ANK
 
 | | orally active thrombin inhibitors in complex with thrombin and an exosite decapeptide | | Descriptor: | N-[(1R)-2-[(1-{[({6-[AMINO(IMINO)METHYL]PYRIDIN-3-YL}METHYL)AMINO]CARBONYL}CYCLOPENTYL)AMINO]-1-(CYCLOHEXYLMETHYL)-2-OXOETHYL]GLYCINE, Thrombin heavy chain, Thrombin light chain, ... | | Authors: | Lange, U.E.W, Baucke, D, Hornberger, W, Mack, H, Seitz, W, Hoeffken, H.W. | | Deposit date: | 2005-08-11 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Orally active thrombin inhibitors. Part 2: optimization of the P2-moiety
BIOORG.MED.CHEM.LETT., 16, 2006
|
|
2AR5
 
 | |
3U13
 
 | | Crystal Structure of de Novo design of cystein esterase ECH13 at the resolution 1.6A, Northeast Structural Genomics Consortium Target OR51 | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Kohan, E, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-29 | | Release date: | 2011-11-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
3U1O
 
 | | THREE DIMENSIONAL STRUCTURE OF DE NOVO DESIGNED CYSTEINE ESTERASE ECH19, Northeast Structural Genomics Consortium Target OR49 | | Descriptor: | De Novo design cysteine esterase ECH19, SODIUM ION, SULFATE ION | | Authors: | Kuzin, A, Su, M, Lew, S, Forouhar, F, Seetharaman, J, Daya, P, Xiao, R, Ciccosanti, C, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-30 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
3U3A
 
 | |
3U6X
 
 | | Phage TP901-1 baseplate tripod | | Descriptor: | BPP, BROMIDE ION, ORF48 | | Authors: | Veesler, D, Spinelli, S, Mahony, J, Lichiere, J, Blangy, S, Bricogne, G, Legrand, P, Ortiz-Lombardia, M, Campanacci, V.I, van Sinderen, D, Cambillau, C. | | Deposit date: | 2011-10-13 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6ZF6
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | 1-[3-[2-hydroxy-2-oxoethyl(phenylsulfonyl)amino]phenyl]-5-[(1~{S},2~{S})-2-phenylcyclopropyl]pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6ZF5
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | 1-[3-[(4-butylphenyl)sulfonyl-(2-hydroxy-2-oxoethyl)amino]phenyl]-5-cyclopropyl-pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6ZEY
 
 | |
6ZEW
 
 | | Keap1 kelch domain bound to a small molecule fragment | | Descriptor: | 7-methoxy-1~{H}-benzotriazole, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6ZEX
 
 | | Keap1 kelch domain bound to a small molecule fragment | | Descriptor: | 5-cyclopropyl-1-phenyl-pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1 | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6ZF1
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | 5-cyclopropyl-1-[3-[2-hydroxy-2-oxoethyl(phenylsulfonyl)amino]phenyl]pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6ZF4
 
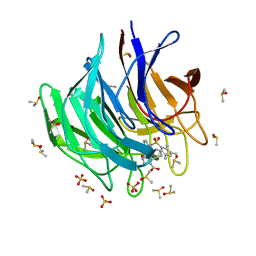 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | 5-cyclopropyl-1-[3-[2-hydroxy-2-oxoethyl-(2,3,5,6-tetramethylphenyl)sulfonyl-amino]phenyl]pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6ZF2
 
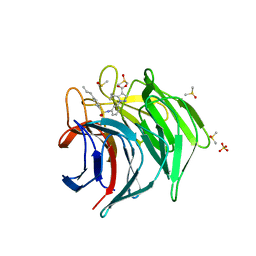 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | 5-cyclopropyl-1-[3-[(4-propylphenyl)sulfonylamino]phenyl]pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6ZF7
 
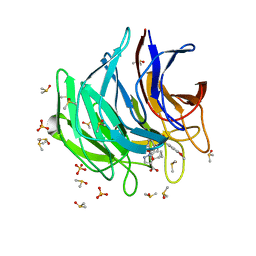 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | 5-cyclopropyl-1-[3-[2-hydroxy-2-oxoethyl-(3-methoxyphenyl)sulfonyl-amino]phenyl]pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6ZF3
 
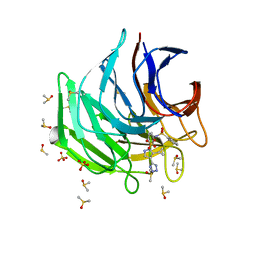 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, SULFATE ION, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6ZF0
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | 5-cyclopropyl-1-[3-[(2,3,5,6-tetramethylphenyl)sulfonylamino]phenyl]pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6TSL
 
 | | Marasmius oreades agglutinin (MOA) in complex with the truncated PVPRAHS synthetic substrate | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin, CALCIUM ION, ... | | Authors: | Cordara, G, Manna, D, Krengel, U. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of MOA in complex with a peptide fragment: A protease caught in flagranti .
Curr Res Struct Biol, 2, 2020
|
|
6TSZ
 
 | | The ULK4 Pseudokinase Domain Bound To ATPgammaS | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase ULK4 | | Authors: | Preuss, F, Chatterjee, D, Mathea, S, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Knapp, S. | | Deposit date: | 2019-12-22 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleotide Binding, Evolutionary Insights, and Interaction Partners of the Pseudokinase Unc-51-like Kinase 4.
Structure, 28, 2020
|
|
6TTT
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 2 (ASI_M3M_140) | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-~{N}-methyl-3,4-bis(oxidanyl)oxolane-2-carboxamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer.
Chemmedchem, 15, 2020
|
|
