5HD4
 
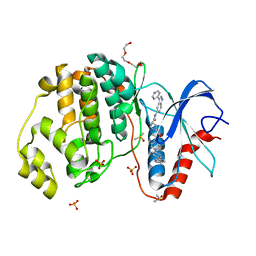 | | Dissecting Therapeutic Resistance to ERK Inhibition Rat Wild Type SCH772984 in complex with (3R)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)-N-[3-(pyridin-4-yl)-2H-indazol-5-yl]pyrrolidine-3-carboxamide | | Descriptor: | (3R)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)-N-[3-(pyridin-4-yl)-2H-indazol-5-yl]pyrrolidine-3-carboxamide, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase 1, ... | | Authors: | Jha, S, Morris, E.J, Hruza, A, Mansueto, M.S, Schroeder, G, Arbanas, J, McMasters, D, Restaino, C.R, Dayananth, R, Black, S, Elsen, N.L, Mannarino, A, Cooper, A, Fawell, S, Zawel, L, Jayaraman, L, Samatar, A.A. | | Deposit date: | 2016-01-04 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Dissecting Therapeutic Resistance to ERK Inhibition.
Mol.Cancer Ther., 15, 2016
|
|
1DJM
 
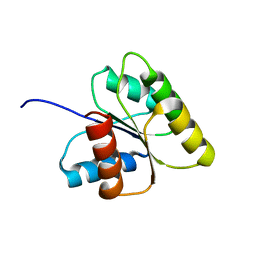 | | SOLUTION STRUCTURE OF BEF3-ACTIVATED CHEY FROM ESCHERICHIA COLI | | Descriptor: | CHEMOTAXIS PROTEIN Y | | Authors: | Cho, H.S, Lee, S.Y, Yan, D, Pan, X, Parkinson, J.S, Kustu, S, Wemmer, D.E, Pelton, J.G. | | Deposit date: | 1999-12-03 | | Release date: | 2000-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of activated CheY.
J.Mol.Biol., 297, 2000
|
|
4QX0
 
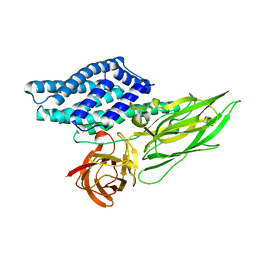 | | Cry3A Toxin structure obtained by Serial Femtosecond Crystallography from in vivo grown crystals isolated from Bacillus thuringiensis and data processed with the cctbx.xfel software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5HS3
 
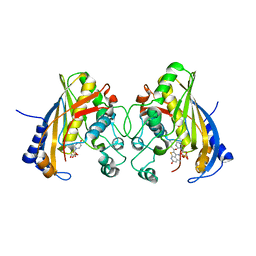 | | Human thymidylate synthase complexed with dUMP and 3-amino-2-benzoyl-4-methylthieno[2,3-b]pyridin-6-ol | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 3-amino-2-benzoyl-4-methylthieno[2,3-b]pyridin-6-ol, Thymidylate synthase | | Authors: | Chen, D, Almqvist, H, Axelsson, H, Jafari, R, Mateus, A, Haraldsson, M, Larsson, A, Artursson, P, Molina, D.M, Lundback, T, Nordlund, P. | | Deposit date: | 2016-01-25 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | CETSA screening identifies known and novel thymidylate synthase inhibitors and slow intracellular activation of 5-fluorouracil
Nat Commun, 7, 2016
|
|
5HVA
 
 | | Crystal structure of DR2231 in complex with dUMPNPP and magnesium. | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DR2231, MAGNESIUM ION | | Authors: | Mota, C.S, Goncalves, A.M.D, de Sanctis, D. | | Deposit date: | 2016-01-28 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Deinococcus radiodurans DR2231 is a two-metal-ion mechanism hydrolase with exclusive activity on dUTP.
FEBS J., 283, 2016
|
|
3PWZ
 
 | | Crystal structure of an Ael1 enzyme from Pseudomonas putida | | Descriptor: | Shikimate dehydrogenase 3 | | Authors: | Christendat, D, Peek, J. | | Deposit date: | 2010-12-09 | | Release date: | 2011-09-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Structural and mechanistic analysis of a novel class of shikimate dehydrogenases: evidence for a conserved catalytic mechanism in the shikimate dehydrogenase family.
Biochemistry, 50, 2011
|
|
3Q2H
 
 | | Adamts1 in complex with N-hydroxyformamide inhibitors of ADAM-TS4 | | Descriptor: | A disintegrin and metalloproteinase with thrombospondin motifs 1, CADMIUM ION, MAGNESIUM ION, ... | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2010-12-20 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The design and synthesis of novel N-hydroxyformamide inhibitors of ADAM-TS4 for the treatment of osteoarthritis
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5HCN
 
 | | GPN-loop GTPase Npa3 in complex with GMPPCP | | Descriptor: | GLYCEROL, GPN-loop GTPase 1, LAURIC ACID, ... | | Authors: | Niesser, J, Wagner, F.R, Kostrewa, D, Muehlbacher, W, Cramer, P. | | Deposit date: | 2016-01-04 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of GPN-Loop GTPase Npa3 and Implications for RNA Polymerase II Assembly.
Mol.Cell.Biol., 36, 2015
|
|
2H8W
 
 | | Solution structure of ribosomal protein L11 | | Descriptor: | 50S ribosomal protein L11 | | Authors: | Lee, D, Walsh, J.D, Yu, P, Choli-Papadopoulou, T, Krueger, S, Draper, D, Wang, Y.-X. | | Deposit date: | 2006-06-08 | | Release date: | 2007-02-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Structure of Free L11 and Functional Dynamics of L11 in Free, L11-rRNA(58 nt) Binary and L11-rRNA(58 nt)-thiostrepton Ternary Complexes.
J.Mol.Biol., 367, 2007
|
|
5HYL
 
 | | Crystal structure of DR2231_E47A mutant in complex with dUMPNPP and magnesium | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DR2231, MAGNESIUM ION | | Authors: | Mota, C.S, Goncalves, A.M.D, de Sanctis, D. | | Deposit date: | 2016-02-01 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Deinococcus radiodurans DR2231 is a two-metal-ion mechanism hydrolase with exclusive activity on dUTP.
FEBS J., 283, 2016
|
|
3U47
 
 | | Human Carbonic Anhydrase II V143L | | Descriptor: | Carbonic anhydrase 2, GLYCEROL, ZINC ION | | Authors: | West, D, Mckenna, R. | | Deposit date: | 2011-10-07 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural modification of the hydrophobic pocket of Human Carbonic Anhydrase II
To be Published
|
|
3PSJ
 
 | |
5HKV
 
 | | The crystal structure of the large ribosomal subunit of Staphylococcus aureus in complex with lincomycin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23s ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Yonath, A, Matzov, D, Eyal, Z, Ben Hamou, R, Zimmerman, E, Rozenberg, H, Bashan, A, Fridman, M. | | Deposit date: | 2016-01-14 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.66 Å) | | Cite: | Structural insights of lincosamides targeting the ribosome of Staphylococcus aureus.
Nucleic Acids Res., 45, 2017
|
|
5HIN
 
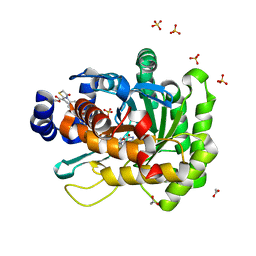 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 18L compound | | Descriptor: | ACETATE ION, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Huang, J, Wu, D, Lu, Q, Yao, X. | | Deposit date: | 2016-01-12 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 18L at 1.60 A resolution
To Be Published
|
|
3PUT
 
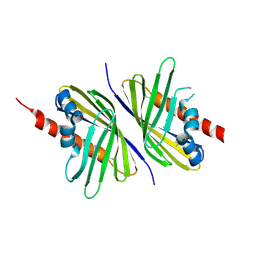 | | Crystal Structure of the CERT START domain (mutant V151E) from Rhizobium etli at the resolution 1.8A, Northeast Structural Genomics Consortium Target ReR239. | | Descriptor: | HEXANE-1,6-DIOL, Hypothetical conserved protein | | Authors: | Kuzin, A, Chen, Y, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-12-06 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.828 Å) | | Cite: | Crystal Structure of the CERT START domain (mutant V151E) from Rhizobium etli at the resolution 1.8A, Northeast Structural Genomics Consortium Target ReR239.
To be Published
|
|
1DUO
 
 | |
3PRU
 
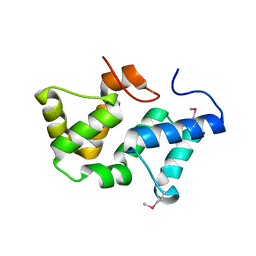 | | Crystal Structure of Phycobilisome 32.1 kDa linker polypeptide, phycocyanin-associated, rod 1 (fragment 14-158) from Synechocystis sp. PCC 6803, Northeast Structural Genomics Consortium Target SgR182A | | Descriptor: | CHLORIDE ION, Phycobilisome 32.1 kDa linker polypeptide, phycocyanin-associated, ... | | Authors: | Kuzin, A, Su, M, Patel, P, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-11-30 | | Release date: | 2010-12-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.677 Å) | | Cite: | Northeast Structural Genomics Consortium Target SgR182A
To be Published
|
|
3PSI
 
 | |
1BHS
 
 | | HUMAN ESTROGENIC 17BETA-HYDROXYSTEROID DEHYDROGENASE | | Descriptor: | 17BETA-HYDROXYSTEROID DEHYDROGENASE | | Authors: | Ghosh, D. | | Deposit date: | 1995-04-19 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human estrogenic 17 beta-hydroxysteroid dehydrogenase at 2.20 A resolution.
Structure, 3, 1995
|
|
3PSF
 
 | |
1B4X
 
 | | ASPARTATE AMINOTRANSFERASE FROM E. COLI, C191S MUTATION, WITH BOUND MALEATE | | Descriptor: | ASPARTATE AMINOTRANSFERASE, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jeffery, C.J, Gloss, L.M, Petsko, G.A, Ringe, D. | | Deposit date: | 1998-12-30 | | Release date: | 2000-10-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The role of residues outside the active site: structural basis for function of C191 mutants of Escherichia coli aspartate aminotransferase.
Protein Eng., 13, 2000
|
|
3SGR
 
 | | Tandem repeat of amyloid-related segment of alphaB-crystallin residues 90-100 mutant V91L | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Tandem repeat of amyloid-related segment of alphaB-crystallin residues 90-100 mutant V91L | | Authors: | Laganowsky, A, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2011-06-15 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Atomic view of a toxic amyloid small oligomer.
Science, 335, 2012
|
|
3SIH
 
 | |
1BBD
 
 | |
1DVH
 
 | | STRUCTURE AND DYNAMICS OF FERROCYTOCHROME C553 FROM DESULFOVIBRIO VULGARIS STUDIED BY NMR SPECTROSCOPY AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | CYTOCHROME C553, HEME C | | Authors: | Blackledge, M.J, Medvedeva, S, Poncin, M, Guerlesquin, F, Bruschi, M, Marion, D. | | Deposit date: | 1995-02-24 | | Release date: | 1995-06-03 | | Last modified: | 2021-03-03 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of ferrocytochrome c553 from Desulfovibrio vulgaris studied by NMR spectroscopy and restrained molecular dynamics.
J.Mol.Biol., 245, 1995
|
|
