3BRH
 
 | |
1FZT
 
 | | SOLUTION STRUCTURE AND DYNAMICS OF AN OPEN B-SHEET, GLYCOLYTIC ENZYME-MONOMERIC 23.7 KDA PHOSPHOGLYCERATE MUTASE FROM SCHIZOSACCHAROMYCES POMBE | | Descriptor: | PHOSPHOGLYCERATE MUTASE | | Authors: | Uhrinova, S, Uhrin, D, Nairn, J, Price, N.C, Fothergill-Gilmore, L.A. | | Deposit date: | 2000-10-04 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of an open beta-sheet, glycolytic enzyme, monomeric 23.7 kDa phosphoglycerate mutase from Schizosaccharomyces pombe.
J.Mol.Biol., 306, 2001
|
|
3BW7
 
 | |
3C0P
 
 | |
1T9I
 
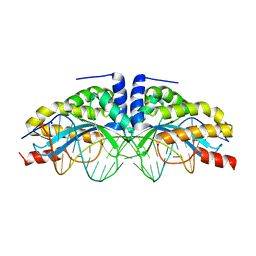 | | I-CreI(D20N)/DNA complex | | Descriptor: | 5'-D(*CP*GP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*TP*TP*TP*GP*C)-3', 5'-D(*GP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*TP*CP*G)-3', CALCIUM ION, ... | | Authors: | Chevalier, B, Sussman, D, Otis, C, Boudreau, D, Turmel, M, Lemieux, C, Stephens, K, Monnat Jr, R.J, Stoddard, B.L. | | Deposit date: | 2004-05-17 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Metal-Dependent DNA Cleavage Mechanism of the I-CreI LAGLIDADG Homing Endonuclease.
Biochemistry, 43, 2004
|
|
3C9J
 
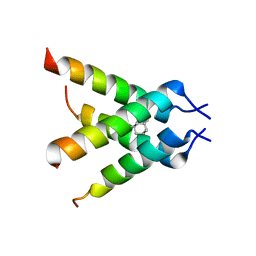 | | The Crystal structure of Transmembrane domain of M2 protein and Amantadine complex | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, Proton Channel protein M2, transmembrane segment | | Authors: | Stouffer, A.L, Acharya, R, Salom, D. | | Deposit date: | 2008-02-15 | | Release date: | 2008-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for the function and inhibition of an influenza virus proton channel
Nature, 451, 2008
|
|
4RW5
 
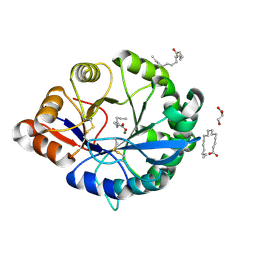 | | Structural insights into substrate binding of brown spider venom class II phospholipases D | | Descriptor: | GLYCEROL, MAGNESIUM ION, N-TRIDECANOIC ACID, ... | | Authors: | Coronado, M.A, Ullah, A, da Silva, L.S, Chaves-Moreira, D, Vuitika, L, Chaim, O.M, Veiga, S.S, Chahine, J, Murakami, M.T, Arni, R.K. | | Deposit date: | 2014-12-01 | | Release date: | 2015-06-03 | | Last modified: | 2015-12-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Insights into Substrate Binding of Brown Spider Venom Class II Phospholipases D.
Curr Protein Pept Sci, 16, 2015
|
|
3AVV
 
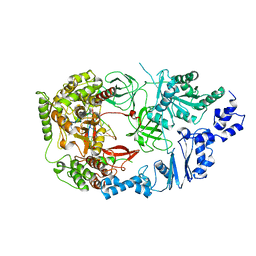 | | Structure of viral RNA polymerase complex 3 | | Descriptor: | CALCIUM ION, Elongation factor Ts, Elongation factor Tu, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2011-03-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.119 Å) | | Cite: | Molecular basis for RNA polymerization by Q beta replicase
Nat.Struct.Mol.Biol., 19, 2012
|
|
3AVY
 
 | | Structure of viral RNA polymerase complex 6 | | Descriptor: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, Elongation factor Ts, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2011-03-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.616 Å) | | Cite: | Molecular basis for RNA polymerization by Q beta replicase
Nat.Struct.Mol.Biol., 19, 2012
|
|
1T9J
 
 | | I-CreI(Q47E)/DNA complex | | Descriptor: | 5'-D(*CP*GP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*TP*TP*TP*GP*C)-3', 5'-D(*GP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*TP*CP*G)-3', DNA endonuclease I-CreI | | Authors: | Chevalier, B, Sussman, D, Otis, C, Boudreau, D, Turmel, M, Lemieux, C, Stephens, K, Monnat Jr, R.J, Stoddard, B.L. | | Deposit date: | 2004-05-17 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal-Dependent DNA Cleavage Mechanism of the I-CreI LAGLIDADG Homing Endonuclease.
Biochemistry, 43, 2004
|
|
4RML
 
 | | Crystal structure of the Olfactomedin domain of latrophilin 3 in C2221 crystal form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Latrophilin-3, MAGNESIUM ION | | Authors: | Ranaivoson, F.M, Liu, Q, Martini, F, Bergami, F, Von daake, S, Li, S, Demeler, B, Hendrickson, W.A, Comoletti, D. | | Deposit date: | 2014-10-21 | | Release date: | 2015-08-19 | | Last modified: | 2015-10-07 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and Mechanistic Insights into the Latrophilin3-FLRT3 Complex that Mediates Glutamatergic Synapse Development.
Structure, 23, 2015
|
|
3ATR
 
 | | Geranylgeranyl Reductase (GGR) from Sulfolobus acidocaldarius co-crystallized with its ligand | | Descriptor: | Conserved Archaeal protein, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, PYROPHOSPHATE, ... | | Authors: | Fujihashi, M, Sasaki, D, Iwata, Y, Yoshimura, T, Hemmi, H, Miki, K. | | Deposit date: | 2011-01-12 | | Release date: | 2011-05-04 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mutation analysis of archaeal geranylgeranyl reductase
J.Mol.Biol., 409, 2011
|
|
3AQC
 
 | | M. luteus B-P 26 heterodimeric hexaprenyl diphosphate synthase in complex with magnesium and FPP analogue | | Descriptor: | (2E,6E)-7,11-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, CHLORIDE ION, Component A of hexaprenyl diphosphate synthase, ... | | Authors: | Sasaki, D, Fujihashi, M, Okuyama, N, Kobayashi, Y, Noike, M, Koyama, T, Miki, K. | | Deposit date: | 2010-10-28 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of heterodimeric hexaprenyl diphosphate synthase from Micrococcus luteus B-P 26 reveals that the small subunit is directly involved in the product chain length regulation.
J.Biol.Chem., 286, 2011
|
|
3AQQ
 
 | | Crystal structure of human CRHSP-24 | | Descriptor: | Calcium-regulated heat stable protein 1 | | Authors: | Hou, H, Wang, F, Zhang, W, Wang, D, Li, X, Bartlam, M, Yao, X, Rao, Z. | | Deposit date: | 2010-11-17 | | Release date: | 2010-12-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | CRHSP-24 is a novel cargo adaptor trafficking between stress granules and processing bodies
To be Published
|
|
4NC3
 
 | | Crystal structure of the 5-HT2B receptor solved using serial femtosecond crystallography in lipidic cubic phase. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHOLESTEROL, ... | | Authors: | Liu, W, Wacker, D, Gati, C, Han, G.W, James, D, Wang, D, Nelson, G, Weierstall, U, Katritch, V, Barty, A, Zatsepin, N.A, Li, D, Messerschmidt, M, Boutet, S, Williams, G.J, Koglin, J.E, Seibert, M.M, Wang, C, Shah, S.T.A, Basu, S, Fromme, R, Kupitz, C, Rendek, K.N, Grotjohann, I, Fromme, P, Kirian, R.A, Beyerlein, K.R, White, T.A, Chapman, H.N, Caffrey, M, Spence, J.C.H, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Serial femtosecond crystallography of G protein-coupled receptors.
Science, 342, 2013
|
|
3AQM
 
 | | Structure of bacterial protein (form II) | | Descriptor: | MAGNESIUM ION, Poly(A) polymerase | | Authors: | Toh, Y, Takeshita, D, Tomita , K. | | Deposit date: | 2010-11-09 | | Release date: | 2011-02-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Mechanism for the alteration of the substrate specificities of template-independent RNA polymerases
Structure, 19, 2011
|
|
1HDC
 
 | |
3AXG
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase | | Descriptor: | Endotype 6-aminohexanoat-oligomer hydrolase, SODIUM ION | | Authors: | Negoro, S, Shibata, N, Tanaka, Y, Yasuhira, K, Shibata, H, Hashimoto, H, Lee, Y.H, Ohshima, S, Santa, R, Mochiji, K, Goto, Y, Ikegami, T, Nagai, K, Kato, D, Takeo, M, Higuchi, Y. | | Deposit date: | 2011-04-04 | | Release date: | 2011-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of nylon hydrolase and mechanism of nylon-6 hydrolysis
J.Biol.Chem., 287, 2012
|
|
3AWR
 
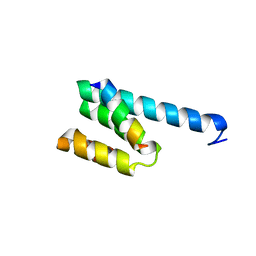 | |
3PA9
 
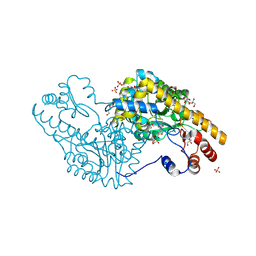 | | Mechanism of inactivation of E. coli aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-furancarboxylic acid (S-ADFA) pH 7.5 | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminofuran-2-carboxylic acid, Aspartate aminotransferase, ... | | Authors: | Liu, D, Pozharski, E, Fu, M, Silverman, R.B, Ringe, D. | | Deposit date: | 2010-10-19 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of inactivation of Escherichia coli aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-furancarboxylic acid .
Biochemistry, 49, 2010
|
|
1UOQ
 
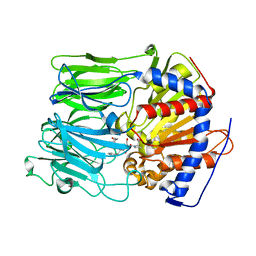 | |
3C8B
 
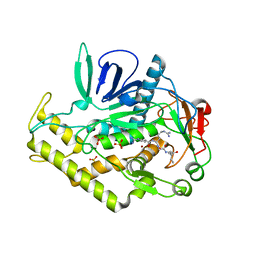 | |
3C89
 
 | |
3C94
 
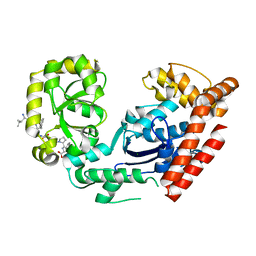 | | ExoI/SSB-Ct complex | | Descriptor: | Exodeoxyribonuclease I, MAGNESIUM ION, Single-stranded DNA-binding C-terminal tail peptide | | Authors: | Lu, D, Keck, J.L. | | Deposit date: | 2008-02-15 | | Release date: | 2008-07-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of Escherichia coli single-stranded DNA-binding protein stimulation of exonuclease I.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3C9F
 
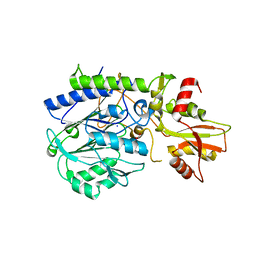 | | Crystal structure of 5'-nucleotidase from Candida albicans SC5314 | | Descriptor: | 5'-nucleotidase, FORMIC ACID, SODIUM ION, ... | | Authors: | Patskovsky, Y, Romero, R, Gilmore, M, Eberle, M, Bain, K, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-15 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of 5'-nucleotidase from Candida albicans.
To be Published
|
|
