4DPF
 
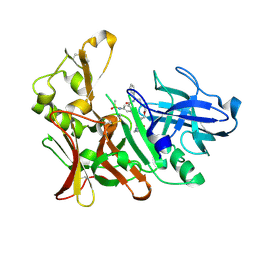 | | BACE-1 in complex with a HEA-macrocyclic type inhibitor | | Descriptor: | Beta-secretase 1, N-[(4S,8E,11S)-4-[(1R)-1-hydroxy-2-{[3-(propan-2-yl)benzyl]amino}ethyl]-2,13-dioxo-11-phenyl-6-oxa-3,12-diazabicyclo[12.3.1]octadeca-1(18),8,14,16-tetraen-16-yl]-N-methylmethanesulfonamide | | Authors: | Lindberg, J, Borkakoti, N, Derbyshire, D. | | Deposit date: | 2012-02-13 | | Release date: | 2012-07-11 | | Last modified: | 2013-01-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Highly potent macrocyclic BACE-1 inhibitors incorporating a hydroxyethylamine core: design, synthesis and X-ray crystal structures of enzyme inhibitor complexes.
Bioorg.Med.Chem., 20, 2012
|
|
4GAG
 
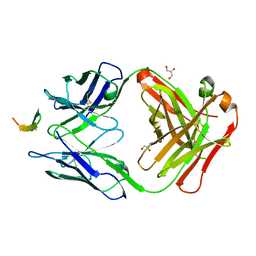 | | Structure of the broadly neutralizing antibody AP33 in complex with its HCV epitope (E2 residues 412-423) | | Descriptor: | GLYCEROL, Genome polyprotein, NEUTRALIZING ANTIBODY AP33 HEAVY CHAIN, ... | | Authors: | Potter, J.A, Owsianka, A, Angus, A.G.N, Jeffery, N, Matthews, D, Keck, Z, Lau, P, Foung, S.K.H, Taylor, G.L, Patel, A.H. | | Deposit date: | 2012-07-25 | | Release date: | 2012-10-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward a Hepatitis C Virus Vaccine: the Structural Basis of Hepatitis C Virus Neutralization by AP33, a Broadly Neutralizing Antibody.
J.Virol., 86, 2012
|
|
4GMI
 
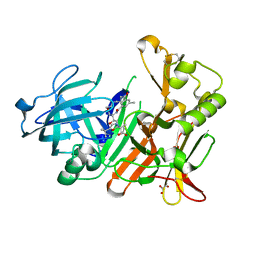 | | BACE-1 in complex with HEA-type macrocyclic inhibitor, MV078571 | | Descriptor: | (4S,8E)-4-[(1R)-2-{[2-(5-tert-butyl-1,3-oxazol-2-yl)propan-2-yl]amino}-1-hydroxyethyl]-16-methyl-6-oxa-3-azabicyclo[12.3.1]octadeca-1(18),8,14,16-tetraene-2,13-dione, ACETATE ION, Beta-secretase 1, ... | | Authors: | Lindberg, J.D, Derbyshire, D. | | Deposit date: | 2012-08-16 | | Release date: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and synthesis of novel BACE-1 inhibitors
To be Published
|
|
4CRM
 
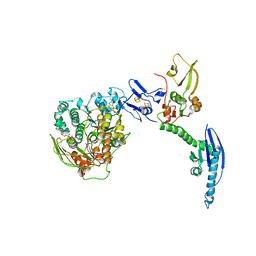 | | Cryo-EM of a pre-recycling complex with eRF1 and ABCE1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, EUKARYOTIC PEPTIDE CHAIN RELEASE FACTOR SUBUNIT 1, ... | | Authors: | Preis, A, Heuer, A, Barrio-Garcia, C, Hauser, A, Eyler, D, Berninghausen, O, Green, R, Becker, T, Beckmann, R. | | Deposit date: | 2014-02-28 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.75 Å) | | Cite: | Cryoelectron Microscopic Structures of Eukaryotic Translation Termination Complexes Containing Erf1-Erf3 or Erf1-Abce1.
Cell Rep., 8, 2014
|
|
4CYF
 
 | | The structure of vanin-1: defining the link between metabolic disease, oxidative stress and inflammation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PANTETHEINASE | | Authors: | Boersma, Y.L, Newman, J, Adams, T.E, Sparrow, L, Cowieson, N, Lucent, D, Krippner, G, Bozaoglu, K, Peat, T.S. | | Deposit date: | 2014-04-11 | | Release date: | 2014-12-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Structure of Vanin-1: A Key Enzyme Linking Metabolic Disease and Inflammation
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4DUL
 
 | | ANAC019 NAC domain crystal form IV | | Descriptor: | NAC domain-containing protein 19 | | Authors: | Lo Leggio, L, Helgstrand, C, Welner, D, Olsen, A.N, Skriver, K. | | Deposit date: | 2012-02-22 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | DNA binding by the plant-specific NAC transcription factors in crystal and solution: a firm link to WRKY and GCM transcription factors.
Biochem.J., 444, 2012
|
|
4I8S
 
 | |
4BGG
 
 | | Crystal structure of the ACVR1 kinase in complex with LDN-213844 | | Descriptor: | 1,2-ETHANEDIOL, 1-{4-[5-(3,4,5-trimethoxyphenyl)pyridin-3-yl]phenyl}piperazine, ACTIVIN RECEPTOR TYPE-1, ... | | Authors: | Sanvitale, C, Canning, P, Cooper, C, Wang, Y, Mohedas, A.H, Choi, S, Yu, P.B, Cuny, G.D, Nowak, R, Coutandin, D, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-26 | | Release date: | 2013-04-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure-Activity Relationship of 3,5-Diaryl-2-Aminopyridine Alk2 Inhibitors Reveals Unaltered Binding Affinity for Fibrodysplasia Ossificans Progressiva Causing Mutants.
J.Med.Chem., 57, 2014
|
|
4E96
 
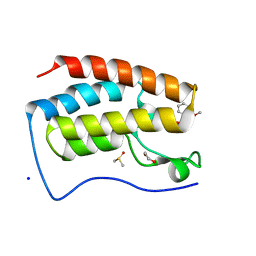 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor PFi-1 | | Descriptor: | 1,2-ETHANEDIOL, 2-methoxy-N-(3-methyl-2-oxo-1,2,3,4-tetrahydroquinazolin-6-yl)benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Fish, P, Bunnage, M, Owen, D, Knapp, S, Cook, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-03-20 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Identification of a chemical probe for bromo and extra C-terminal bromodomain inhibition through optimization of a fragment-derived hit.
J.Med.Chem., 55, 2012
|
|
4CEH
 
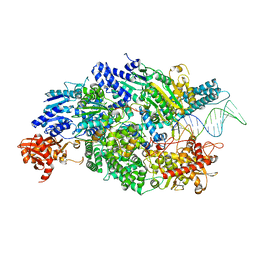 | | Crystal structure of AddAB with a forked DNA substrate | | Descriptor: | ATP-DEPENDENT HELICASE/DEOXYRIBONUCLEASE SUBUNIT B, ATP-DEPENDENT HELICASE/NUCLEASE SUBUNIT A, DNA, ... | | Authors: | Krajewski, W.W, Wilkinson, M, Fu, X, Cronin, N.B, Wigley, D. | | Deposit date: | 2013-11-11 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Structural Basis for Translocation by Addab Helicase-Nuclease and its Arrest at Chi Sites.
Nature, 508, 2014
|
|
4J12
 
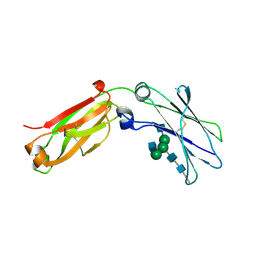 | | monomeric Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, human Fc fragment | | Authors: | Ishino, T, Wang, M, Mosyak, L, Tam, A, Duan, W, Svenson, K, Joyce, A, O'Hara, D, Lin, L, Somers, W, Kriz, R. | | Deposit date: | 2013-01-31 | | Release date: | 2013-05-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineering a Monomeric Fc Domain Modality by N-Glycosylation for the Half-life Extension of Biotherapeutics.
J.Biol.Chem., 288, 2013
|
|
4HYJ
 
 | | Crystal structure of Exiguobacterium sibiricum rhodopsin | | Descriptor: | EICOSANE, RETINAL, Rhodopsin | | Authors: | Gushchin, I, Chervakov, P, Kuzmichev, P, Popov, A, Round, E, Borshchevskiy, V, Dolgikh, D, Kirpichnikov, M, Petrovskaya, L, Chupin, V, Arseniev, A, Gordeliy, V. | | Deposit date: | 2012-11-13 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the proton pumping by unusual proteorhodopsin from nonmarine bacteria.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4CEI
 
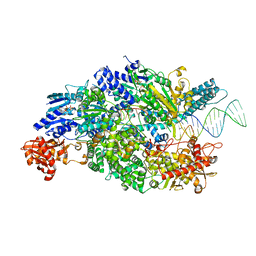 | | Crystal structure of ADPNP-bound AddAB with a forked DNA substrate | | Descriptor: | ATP-DEPENDENT HELICASE/DEOXYRIBONUCLEASE SUBUNIT B, ATP-DEPENDENT HELICASE/NUCLEASE SUBUNIT A, DNA, ... | | Authors: | Krajewski, W.W, Wilkinson, M, Fu, X, Cronin, N.B, Wigley, D. | | Deposit date: | 2013-11-11 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Translocation by Addab Helicase-Nuclease and its Arrest at Chi Sites.
Nature, 508, 2014
|
|
4CEJ
 
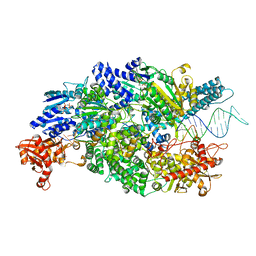 | | Crystal structure of AddAB-DNA-ADPNP complex at 3 Angstrom resolution | | Descriptor: | ATP-DEPENDENT HELICASE/DEOXYRIBONUCLEASE SUBUNIT B, ATP-DEPENDENT HELICASE/NUCLEASE SUBUNIT A, DNA, ... | | Authors: | Krajewski, W.W, Wilkinson, M, Fu, X, Cronin, N.B, Wigley, D. | | Deposit date: | 2013-11-11 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Translocation by Addab Helicase-Nuclease and its Arrest at Chi Sites.
Nature, 508, 2014
|
|
4E67
 
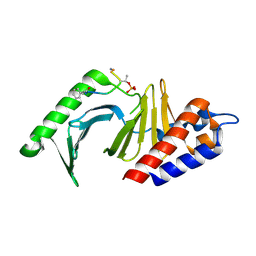 | | The structure of the polo-box domain (PBD) of polo-like kinase 1 (Plk1) in complex with hydrocinnamoyl-derivatized PLHSpTA peptide | | Descriptor: | Serine/threonine-protein kinase PLK1, hydrocinnamoyl-derivatized PLHSpTA peptide | | Authors: | Sledz, P, Hyvonen, M, Tan, Y.S, Lang, S, Spring, D, Abell, C, Best, R.B. | | Deposit date: | 2012-03-15 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Using ligand-mapping simulations to design a ligand selectively targeting a cryptic surface pocket of polo-like kinase 1.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4GVU
 
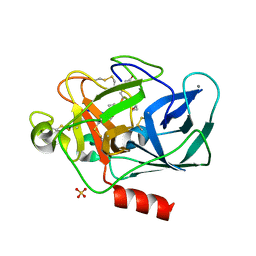 | | Lyngbyastatin 7-Porcine Pancreatic Elastase Co-crystal Structure | | Descriptor: | CALCIUM ION, Chymotrypsin-like elastase family member 1, Lyngbyastatin 7, ... | | Authors: | Salvador, L.A, Taori, K, Biggs, J.S, Jakoncic, J, Ostrov, D, Paul, V.J, Luesch, H. | | Deposit date: | 2012-08-31 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Potent elastase inhibitors from cyanobacteria: structural basis and mechanisms mediating cytoprotective and anti-inflammatory effects in bronchial epithelial cells.
J.Med.Chem., 56, 2013
|
|
4GX9
 
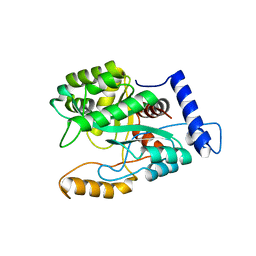 | | Crystal structure of a DNA polymerase III alpha-epsilon chimera | | Descriptor: | DNA polymerase III subunit epsilon,DNA polymerase III subunit alpha | | Authors: | Li, N, Horan, N, Xu, Z.-Q, Jacques, D, Dixon, N.E, Oakley, A.J. | | Deposit date: | 2012-09-04 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Proofreading exonuclease on a tether: the complex between the E. coli DNA polymerase III subunits alpha, {varepsilon}, theta and beta reveals a highly flexible arrangement of the proofreading domain
Nucleic Acids Res., 41, 2013
|
|
4J51
 
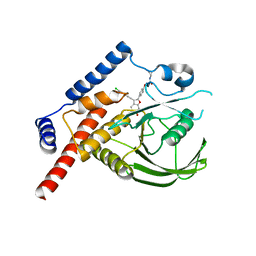 | |
4CRN
 
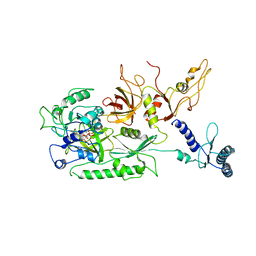 | | Cryo-EM of a pretermination complex with eRF1 and eRF3 | | Descriptor: | ERF1 IN RIBOSOME-BOUND ERF1-ERF3-GDPNP COMPLEX, ERF3 IN RIBOSOME BOUND ERF1-ERF3-GDPNP COMPLEX, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Preis, A, Heuer, A, Barrio-Garcia, C, Hauser, A, Eyler, D, Berninghausen, O, Green, R, Becker, T, Beckmann, R. | | Deposit date: | 2014-02-28 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Cryoelectron Microscopic Structures of Eukaryotic Translation Termination Complexes Containing Erf1-Erf3 or Erf1-Abce1.
Cell Rep., 8, 2014
|
|
4DPI
 
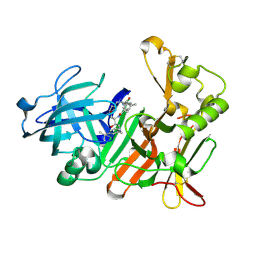 | | BACE-1 in complex with HEA-macrocyclic inhibitor, MV078512 | | Descriptor: | (4S,8E,11R)-4-[(1R)-1-hydroxy-2-{[3-(propan-2-yl)benzyl]amino}ethyl]-16-methyl-11-phenyl-6-oxa-3,12-diazabicyclo[12.3.1]octadeca-1(18),8,14,16-tetraene-2,13-dione, Beta-secretase 1 | | Authors: | Lindberg, J, Borkakoti, N, Derbyshire, D. | | Deposit date: | 2012-02-13 | | Release date: | 2012-07-11 | | Last modified: | 2013-01-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly potent macrocyclic BACE-1 inhibitors incorporating a hydroxyethylamine core: design, synthesis and X-ray crystal structures of enzyme inhibitor complexes.
Bioorg.Med.Chem., 20, 2012
|
|
4H7C
 
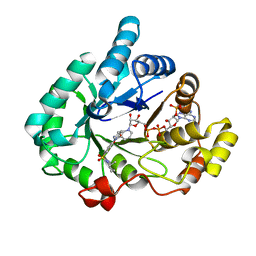 | | Crystal structure of human 17beta-hydroxysteroid dehydrogenase type 5 in complex with 1-{4-[(2-methyl-1-piperidinyl)sulfonyl]phenyl}-2-pyrrolidinone | | Descriptor: | 1-(4-{[(2R)-2-methylpiperidin-1-yl]sulfonyl}phenyl)-1,3-dihydro-2H-pyrrol-2-one, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Turnbull, A.P, Heinrich, D, Jamieson, S.M.F, Flanagan, J.U, Silva, S, Rigoreau, L.J.M, Trivier, E, Soudy, C, Samlal, S.S, Owen, P.J, Schroeder, E, Raynham, T, Denny, W.A. | | Deposit date: | 2012-09-20 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Synthesis and structure-activity relationships for 1-(4-(piperidin-1-ylsulfonyl)phenyl)pyrrolidin-2-ones as novel non-carboxylate inhibitors of the aldo-keto reductase enzyme AKR1C3.
Eur.J.Med.Chem., 62C, 2013
|
|
4ER2
 
 | | The active site of aspartic proteinases | | Descriptor: | ENDOTHIAPEPSIN, PEPSTATIN, SULFATE ION | | Authors: | Bailey, D, Veerapandian, B, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1990-10-20 | | Release date: | 1991-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The active site of aspartic proteinases
FEBS Lett., 174, 1984
|
|
4D6P
 
 | | RADA C-TERMINAL ATPASE DOMAIN FROM PYROCOCCUS FURIOSUS BOUND TO AMPPNP | | Descriptor: | DNA REPAIR AND RECOMBINATION PROTEIN RADA, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Marsh, M.E, Ehebauer, M.T, Scott, D, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2014-11-13 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | ATP Half-Sites in Rada and Rad51 Recombinases Bind Nucleotides
FEBS Open Bio, 6, 2016
|
|
4J2R
 
 | | Middle domain of influenza A virus RNA-dependent polymerase PB2 | | Descriptor: | Polymerase basic protein 2 | | Authors: | Qiu, H, Tsurumura, T, Tsumori, Y, Hatakeyama, D, Kuzuhara, T, Tsuge, H. | | Deposit date: | 2013-02-05 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystallization and preliminary X-ray diffraction studies of a surface mutant of the middle domain of PB2 from human influenza A (H1N1) virus
ACTA CRYSTALLOGR.,SECT.F, 70, 2014
|
|
4J35
 
 | | Molecular Engineering of Organophosphate Hydrolysis Activity from a Weak Promiscuous Lactonase Template | | Descriptor: | COBALT (II) ION, Phosphotriesterase, putative | | Authors: | Sterner, R, Raushel, F, Meier, M, Rajendran, C, Malisi, C, Fox, N, Schlee, S, Barondeau, D, Cker, B.H. | | Deposit date: | 2013-02-05 | | Release date: | 2013-07-24 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Molecular engineering of organophosphate hydrolysis activity from a weak promiscuous lactonase template.
J.Am.Chem.Soc., 135, 2013
|
|
