3OPD
 
 | | Crystal Structure of the N-terminal domain of an HSP90 from Trypanosoma Brucei, Tb10.26.1080 in the presence of a benzamide derivative | | Descriptor: | 4-[6,6-dimethyl-4-oxo-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl]-2-[(cis-4-hydroxycyclohexyl)amino]benzamide, Heat shock protein 83 | | Authors: | Pizarro, J.C, Wernimont, A.K, Hutchinson, A, Sullivan, H, Chamberlain, K, Weadge, J, Cossar, D, Li, Y, Kozieradzki, I, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Wyatt, P.G, Fairlamb, A.H, MacKenzie, C, Ferguson, M.A.J, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-31 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Exploring the Trypanosoma brucei Hsp83 potential as a target for structure guided drug design.
PLoS Negl Trop Dis, 7, 2013
|
|
2JTY
 
 | | Self-complemented variant of FimA, the main subunit of type 1 pilus | | Descriptor: | Type-1 fimbrial protein, A chain | | Authors: | Erilov, D, Wider, G, Glockshuber, R, Puorger, C, Vetsch, M. | | Deposit date: | 2007-08-09 | | Release date: | 2008-08-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure, Folding and Stability of FimA, the Main Structural Subunit of Type 1 Pili from Uropathogenic Escherichia coli Strains.
J.Mol.Biol., 412, 2011
|
|
3P8H
 
 | | Crystal structure of L3MBTL1 (MBT repeat) in complex with a nicotinamide antagonist | | Descriptor: | 3-bromo-5-[(4-pyrrolidin-1-ylpiperidin-1-yl)carbonyl]pyridine, GLYCEROL, Lethal(3)malignant brain tumor-like protein, ... | | Authors: | Lam, R, Herold, J.M, Ouyang, H, Tempel, W, Gao, C, Ravichandran, M, Senisterra, G, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Kireev, D, Frye, S.V, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-13 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Small-molecule ligands of methyl-lysine binding proteins.
J.Med.Chem., 54, 2011
|
|
7EB2
 
 | | Cryo-EM structure of human GABA(B) receptor-Gi protein complex | | Descriptor: | (3S)-5,7-ditert-butyl-3-oxidanyl-3-(trifluoromethyl)-1-benzofuran-2-one, Gamma-aminobutyric acid type B receptor subunit 1, Gamma-aminobutyric acid type B receptor subunit 2, ... | | Authors: | Shen, C, Mao, C, Xu, C, Jin, N, Zhang, H, Shen, D, Shen, Q, Wang, X, Hou, T, Rondard, P, Chen, Z, Pin, J, Zhang, Y, Liu, J. | | Deposit date: | 2021-03-08 | | Release date: | 2021-05-05 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of GABA B receptor-G i protein coupling.
Nature, 594, 2021
|
|
7DZY
 
 | | Spike protein from SARS-CoV2 with Fab fragment of enhancing antibody 2490 | | Descriptor: | Fab Heavy chain of enhancing antibody 2490, Fab light chain of enhancing antibody 2490, Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
7DZW
 
 | | Apo spike protein from SARS-CoV2 | | Descriptor: | Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
7E7X
 
 | |
7DZX
 
 | | Spike protein from SARS-CoV2 with Fab fragment of enhancing antibody 8D2 | | Descriptor: | Fab Heavy chain of enhancing antibody, Fab light chain of enhancing antibody, Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
2GBH
 
 | | NMR structure of stem region of helix-35 of 23S E.coli ribosomal RNA (residues 736-760) | | Descriptor: | 5'-R(*(GMP)P*GP*GP*CP*UP*AP*AP*UP*GP*(PSU)P*UP*GP*AP*AP*AP*AP*AP*UP*UP*AP*GP*CP*CP*C)-3' | | Authors: | Bax, A, Boisbouvier, J, Bryce, D, Grishaev, A, Jaroniec, C, Miclet, E, Nikonovicz, E, O'Neil-Cabello, E, Ying, J. | | Deposit date: | 2006-03-10 | | Release date: | 2006-04-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Measurement of five dipolar couplings from a single 3D NMR multiplet applied to the study of RNA dynamics.
J.Am.Chem.Soc., 126, 2004
|
|
2HEQ
 
 | | NMR Structure of Bacillus subtilis protein YorP, Northeast Structural Genomics Target SR399. | | Descriptor: | YorP protein | | Authors: | Ramelot, T.A, Cort, J.R, Wang, D, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.M, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-21 | | Release date: | 2006-08-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Bacillus subtilis protein YorP, Northeast Structural Genomics Target SR399.
To be Published
|
|
2HC5
 
 | | Solution NMR Structure of Protein yvyC from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR482. | | Descriptor: | Hypothetical protein yvyC | | Authors: | Eletsky, A, Liu, G, Atreya, H.S, Sukumaran, D.K, Wang, D, Cunningham, K, Janjua, H, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-15 | | Release date: | 2006-08-15 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of protein YvyC from Bacillus subtilis reveals unexpected structural similarity between two PFAM families.
Proteins, 76, 2009
|
|
2JUE
 
 | |
2JXH
 
 | |
2JZ5
 
 | | NMR solution structure of protein VPA0419 from Vibrio parahaemolyticus. Northeast Structural Genomics target VpR68 | | Descriptor: | Uncharacterized protein VPA0419 | | Authors: | Singarapu, K.K, Sukumaran, D.K, Eletski, A, Parish, D, Zhao, L, Jiang, M, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-12-28 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structures of proteins VPA0419 from Vibrio parahaemolyticus and yiiS from Shigella flexneri provide structural coverage for protein domain family PFAM 04175.
Proteins, 78, 2010
|
|
2JXT
 
 | | Solution structure of 50S ribosomal protein LX from Methanobacterium thermoautotrophicum. Northeast Structural Genomics Consortium (NESG) target TR80 | | Descriptor: | 50S ribosomal protein LX | | Authors: | Liu, G, Wang, D, Nwosu, C, Owens, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-29 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of 50S ribosomal protein LX from Methanobacterium thermoautotrophicum.
To be Published
|
|
2JXP
 
 | | Solution NMR structure of uncharacterized lipoprotein B from Nitrosomonas europaea. Northeast Structural Genomics target NeR45A | | Descriptor: | Putative lipoprotein B | | Authors: | Rossi, P, Wang, D, Janjua, H, Owens, L, Xiao, R, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-27 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of uncharacterized lipoprotein B from Nitrosomonas europaea.
To be Published
|
|
2I1N
 
 | | Crystal structure of the 1st PDZ domain of Human DLG3 | | Descriptor: | Discs, large homolog 3, SODIUM ION | | Authors: | Turnbull, A.P, Phillips, C, Bunkoczi, G, Debreczeni, J, Ugochukwu, E, Pike, A.C.W, Gorrec, F, Umeano, C, Elkins, J, Berridge, G, Savitsky, P, Gileadi, O, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-14 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions.
Protein Sci., 16, 2007
|
|
3NIO
 
 | | Crystal structure of Pseudomonas aeruginosa guanidinobutyrase | | Descriptor: | Guanidinobutyrase, MANGANESE (II) ION | | Authors: | Lee, S.J, Kim, H.S, Kim, D.J, Yoon, H.J, Kim, K.H, Yoon, J.Y, Jang, J.Y, Im, H, An, D, Suh, S.W. | | Deposit date: | 2010-06-16 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of Pseudomonas aeruginosa guanidinobutyrase and guanidinopropionase, members of the ureohydrolase superfamily
J.Struct.Biol., 175, 2011
|
|
2HVL
 
 | |
2HW4
 
 | | Crystal structure of human phosphohistidine phosphatase | | Descriptor: | 14 kDa phosphohistidine phosphatase, FORMIC ACID | | Authors: | Busam, R.D, Thorsell, A.G, Arrowsmith, C, Berglund, H, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Holmberg Schiavone, L, Hogbom, M, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Ogg, D, Stenmark, P, Sundstrom, M, Uppenberg, J, Van Den Berg, S, Weigelt, J, Persson, C, Hallberg, B.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-07-31 | | Release date: | 2006-08-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | First structure of a eukaryotic phosphohistidine phosphatase
J.Biol.Chem., 281, 2006
|
|
7EHZ
 
 | | Structure of human NNMT in complex with macrocyclic peptide 2 | | Descriptor: | Nicotinamide N-methyltransferase, macrocyclic peptide 2 | | Authors: | Hayashi, K, Mikamiyama, H, Uehara, S, Yamamoto, S, Cary, D, Nishikawa, J, Ueda, T, Ozasa, H, Mihara, K, Yoshimura, N, Kawai, T, Ono, T, Yamamoto, S, Fumoto, M. | | Deposit date: | 2021-03-30 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Macrocyclic Peptides as a Novel Class of NNMT Inhibitors: A SAR Study Aimed at Inhibitory Activity in the Cell.
Acs Med.Chem.Lett., 12, 2021
|
|
7EGU
 
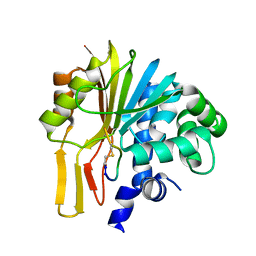 | | Structure of human NNMT in complex with macrocyclic peptide X | | Descriptor: | Nicotinamide N-methyltransferase, macrocyclic peptide X | | Authors: | Hayashi, K, Mikamiyama, H, Uehara, S, Yamamoto, S, Cary, D, Nishikawa, J, Ueda, T, Ozasa, H, Mihara, K, Yoshimura, N, Kawai, T, Ono, T, Yamamoto, S, Fumoto, M. | | Deposit date: | 2021-03-26 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Macrocyclic Peptides as a Novel Class of NNMT Inhibitors: A SAR Study Aimed at Inhibitory Activity in the Cell.
Acs Med.Chem.Lett., 12, 2021
|
|
2FY1
 
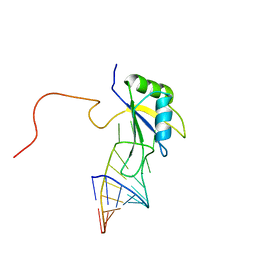 | | A dual mode of RNA recognition by the RBMY protein | | Descriptor: | RNA-binding motif protein, Y chromosome, family 1 member A1, ... | | Authors: | Skrisovska, L, Bourgois, C, Stefl, R, Kister, L, Wenter, P, Elliot, D, Stevenin, J, Allain, F.H.T. | | Deposit date: | 2006-02-07 | | Release date: | 2007-02-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The testis-specific human protein RBMY recognizes RNA through a novel mode of interaction.
EMBO Rep., 8, 2007
|
|
2JXG
 
 | |
2JXI
 
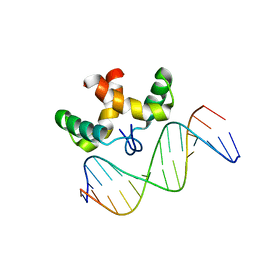 | | Solution structure of the DNA-binding domain of Pseudomonas putida Proline utilization A (putA) bound to GTTGCA DNA sequence | | Descriptor: | DNA (5'-D(*DAP*DAP*DAP*DGP*DGP*DTP*DGP*DCP*DAP*DAP*DCP*DCP*DGP*DC)-3'), DNA (5'-D(*DGP*DCP*DGP*DGP*DTP*DTP*DGP*DCP*DAP*DCP*DCP*DTP*DTP*DT)-3'), Proline dehydrogenase | | Authors: | Halouska, S, Zhou, Y, Becker, D, Powers, R. | | Deposit date: | 2007-11-19 | | Release date: | 2008-10-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Pseudomonas putida protein PpPutA45 and its DNA complex
Proteins, 75, 2008
|
|
