6ZY1
 
 | |
5NK8
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 2f | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[[3-[(2,2,6,6-tetramethyl-1-oxidanyl-piperidin-4-yl)carbamoyl]phenyl]amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5NKG
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 3d | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[[3-ethyl-5-(piperidin-4-ylcarbamoyl)phenyl]amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
8D2X
 
 | | Zebrafish MFSD2A isoform B in inward open ligand 3C conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
3UIN
 
 | | Complex between human RanGAP1-SUMO2, UBC9 and the IR1 domain from RanBP2 | | Descriptor: | E3 SUMO-protein ligase RanBP2, Ran GTPase-activating protein 1, SUMO-conjugating enzyme UBC9, ... | | Authors: | Gareau, J.R, Reverter, D, Lima, C.D. | | Deposit date: | 2011-11-05 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Determinants of small ubiquitin-like modifier 1 (SUMO1) protein specificity, E3 ligase, and SUMO-RanGAP1 binding activities of nucleoporin RanBP2.
J.Biol.Chem., 287, 2012
|
|
6ZQI
 
 | | Cryo-EM structure of Spondweni virus prME | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein | | Authors: | Renner, M, Dejnirattisai, W, Carrique, L, Serna Martin, I, Karia, D, Ilca, S.L, Ho, S.F, Kotecha, A, Keown, J.R, Mongkolsapaya, J, Screaton, G.R, Grimes, J.M. | | Deposit date: | 2020-07-09 | | Release date: | 2021-01-20 | | Last modified: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Flavivirus maturation leads to the formation of an occupied lipid pocket in the surface glycoproteins.
Nat Commun, 12, 2021
|
|
5NMX
 
 | | Crystal Structure of the pyrrolizidine alkaloid N-oxygenase from Zonocerus variegatus in complex with FAD and NADP+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase, MAGNESIUM ION, ... | | Authors: | Scheidig, A, Kubitza, C, Faust, A, Ober, D. | | Deposit date: | 2017-04-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of pyrrolizidine alkaloid N-oxygenase from the grasshopper Zonocerus variegatus.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
8CTN
 
 | | Structure of a K+ selective NaK mutant (NaK2K, Laue diffraction, no electric field) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
6ZY0
 
 | | Catabolic reductive dehalogenase NpRdhA, N-terminally tagged, K488Q variant | | Descriptor: | CHLORIDE ION, COBALAMIN, IRON/SULFUR CLUSTER, ... | | Authors: | Leys, D, Halliwell, T. | | Deposit date: | 2020-07-30 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Catabolic Reductive Dehalogenase Substrate Complex Structures Underpin Rational Repurposing of Substrate Scope.
Microorganisms, 8, 2020
|
|
4RF5
 
 | | Crystal structure of ketoreductase from Lactobacillus kefir, E145S mutant | | Descriptor: | GLYCEROL, MAGNESIUM ION, NADPH dependent R-specific alcohol dehydrogenase | | Authors: | Tang, Y, Tibrewal, N, Cascio, D. | | Deposit date: | 2014-09-24 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.596 Å) | | Cite: | Origins of stereoselectivity in evolved ketoreductases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8CTT
 
 | | Crystal structure of a K+ selective NaK mutant (NaK2K) at 100K | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CTW
 
 | | Crystal structure of a K+ selective NaK mutant (NaK2K) -Na+,Tl+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Potassium channel protein, SODIUM ION, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
5NP0
 
 | | Closed dimer of human ATM (Ataxia telangiectasia mutated) | | Descriptor: | Serine-protein kinase ATM | | Authors: | Baretic, D, Pollard, H.K, Fisher, D.I, Johnson, C.M, Santhanam, B, Truman, C.M, Kouba, T, Fersht, A.R, Phillips, C, Williams, R.L. | | Deposit date: | 2017-04-13 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of closed and open conformations of dimeric human ATM.
Sci Adv, 3, 2017
|
|
7A78
 
 | | Crystal structure of RXR beta LBD in complexes with palmitic acid and GRIP-1 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Nuclear receptor coactivator 2, ... | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Comprehensive Set of Tertiary Complex Structures and Palmitic Acid Binding Provide Molecular Insights into Ligand Design for RXR Isoforms.
Int J Mol Sci, 21, 2020
|
|
8CTX
 
 | | Crystal structure of a K+ selective NaK mutant (NaK2K) -K+,Tl+ complex | | Descriptor: | POTASSIUM ION, Potassium channel protein, THALLIUM (I) ION | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
7AAC
 
 | | Crystal structure of the catalytic domain of human PARP1 in complex with veliparib | | Descriptor: | (2R)-2-(7-carbamoyl-1H-benzimidazol-2-yl)-2-methylpyrrolidinium, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Easton, L.E, Underwood, E, Rawlins, P.B, Johannes, J.W, Embrey, K.J, Neuhaus, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.593 Å) | | Cite: | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
8DE4
 
 | |
5N5S
 
 | | Crystal structure of aldehyde dehydrogenase 21 (ALDH21) from Physcomitrella patens in complex with NADP+ | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde dehydrogenase 21 (ALDH21), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Kopecny, D, Vigouroux, A, Briozzo, P, Morera, S. | | Deposit date: | 2017-02-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The ALDH21 gene found in lower plants and some vascular plants codes for a NADP(+) -dependent succinic semialdehyde dehydrogenase.
Plant J., 92, 2017
|
|
5N6Q
 
 | | Xenobiotic reductase A (XenA) from Pseudomonas putida in complex with 2-phenylacrylic acid | | Descriptor: | 2-Phenylacrylic acid, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Karuppiah, V, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the ene-reductase synthesis of profens.
Org. Biomol. Chem., 15, 2017
|
|
8CTV
 
 | | Crystal structure of a K+ selective NaK mutant (NaK2K) -Tl+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Potassium channel protein, SODIUM ION, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
7A2D
 
 | | Structure-function analyses of dual-BON domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation to the cell division site | | Descriptor: | Uncharacterized protein YraP | | Authors: | Bryant, J.A, Morris, F.C, Knowles, T.J, Maderbocus, R, Heinz, E, Boelter, G, Alodaini, D, Colyer, A, Wotherspoon, P.J, Staunton, K.A, Jeeves, M, Browning, D.F, Sevastsyanovich, Y.R, Wells, T.J, Rossiter, A.E, Bavro, V.N, Sridhar, P, Ward, D.G, Chong, Z.S, Goodall, E.C.A, Icke, C, Teo, A, Chng, S.S, Roper, D.I, Lithgow, T, Cunningham, A.F, Banzhaf, M, Overduin, M, Henderson, I.R. | | Deposit date: | 2020-08-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of dual BON-domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation.
Elife, 9, 2020
|
|
7A77
 
 | | Crystal structure of RXR alpha LBD in complexes with palmitic acid and GRIP-1 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Nuclear receptor coactivator 2, ... | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Comprehensive Set of Tertiary Complex Structures and Palmitic Acid Binding Provide Molecular Insights into Ligand Design for RXR Isoforms.
Int J Mol Sci, 21, 2020
|
|
7A79
 
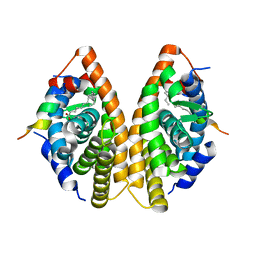 | | Crystal structure of RXR gamma LBD in complexes with palmitic acid and GRIP-1 peptide | | Descriptor: | Nuclear receptor coactivator 2, PALMITIC ACID, Retinoic acid receptor RXR-gamma | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Comprehensive Set of Tertiary Complex Structures and Palmitic Acid Binding Provide Molecular Insights into Ligand Design for RXR Isoforms.
Int J Mol Sci, 21, 2020
|
|
8CTS
 
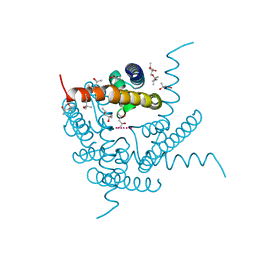 | | Room temperature crystal structure of a K+ selective NaK mutant (NaK2K) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
5N6V
 
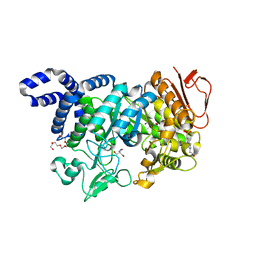 | | Crystal structure of Neisseria polysaccharea amylosucrase mutant derived from Neutral genetic Drift-based engineering | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Amylosucrase, ... | | Authors: | Daude, D, Verges, A, Tranier, S. | | Deposit date: | 2017-02-16 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Neutral Genetic Drift-Based Engineering of a Sucrose-Utilizing Enzyme toward Glycodiversification.
Acs Catalysis, 2019
|
|
