7NHV
 
 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ016 | | Descriptor: | (R)-N-((1-(6-(benzylamino)pyrimidin-4-yl)-3-hydroxypiperidin-3-yl)methyl)-4-((4,4-dimethylpiperidin-1-yl)methyl)benzamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-02-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
7NHH
 
 | |
7NI7
 
 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ031 | | Descriptor: | (R)-4-((4,4-dimethylpiperidin-1-yl)methyl)-2-hydroxy-N-((3-hydroxy-1-(6-((3-(methylcarbamoyl)benzyl)amino)pyrimidin-4-yl)piperidin-3-yl)methyl)benzamide, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-02-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
7NID
 
 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ078 | | Descriptor: | (R)-1-(6-(benzylamino)pyrimidin-4-yl)-3-(((6-((4,4-dimethylpiperidin-1-yl)methyl)naphthalen-1-yl)amino)methyl)piperidin-3-ol, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-02-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
7NHJ
 
 | |
1NB3
 
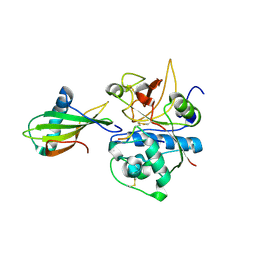 | | Crystal structure of stefin A in complex with cathepsin H: N-terminal residues of inhibitors can adapt to the active sites of endo-and exopeptidases | | Descriptor: | CATHEPSIN H MINI CHAIN, Cathepsin H, Stefin A, ... | | Authors: | Jenko, S, Dolenc, I, Guncar, G, Dobersek, A, Podobnik, M, Turk, D. | | Deposit date: | 2002-12-02 | | Release date: | 2003-02-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of stefin A in complex with cathepsin H: N-terminal residues of inhibitors can adapt to the active sites of endo- and exopeptidases
J.Mol.Biol., 326, 2003
|
|
7NI8
 
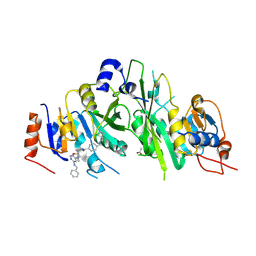 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ040a | | Descriptor: | (R)-4-((2-azaspiro[3.3]heptan-2-yl)methyl)-N-((1-(6-(benzylamino)pyrimidin-4-yl)-3-hydroxypiperidin-3-yl)methyl)benzamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-02-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
1NI0
 
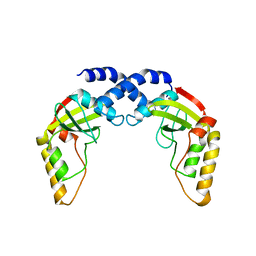 | |
1NIP
 
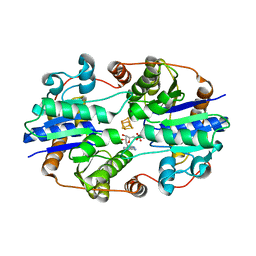 | | CRYSTALLOGRAPHIC STRUCTURE OF THE NITROGENASE IRON PROTEIN FROM AZOTOBACTER VINELANDII | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Komiya, H, Georgiadis, M.M, Chakrabarti, P, Woo, D, Kornuc, J.J, Rees, D.C. | | Deposit date: | 1992-09-29 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii.
Science, 257, 1992
|
|
8A5O
 
 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of S. cerevisiae INO80 | | Descriptor: | Actin, Actin-like protein ARP8, Actin-related protein 4, ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-06-15 | | Release date: | 2022-12-14 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
7N7D
 
 | |
7N7E
 
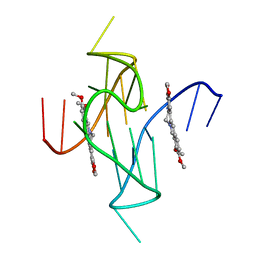 | |
2VFL
 
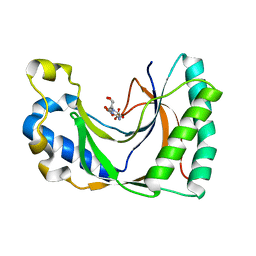 | | AKAP18 delta central domain - CMP | | Descriptor: | AKAP18 DELTA, CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Gold, M.G, Smith, F.D, Scott, J.D, Barford, D. | | Deposit date: | 2007-11-05 | | Release date: | 2008-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Akap18 Contains a Phosphoesterase Domain that Binds AMP
J.Mol.Biol., 375, 2008
|
|
2K3N
 
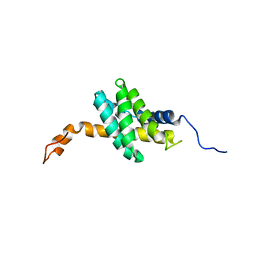 | |
2UV3
 
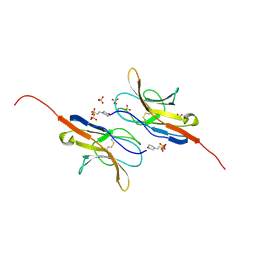 | | Structure of the signal-regulatory protein (SIRP) alpha domain that binds CD47. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE SUBSTRATE 1 | | Authors: | Hatherley, D, Harlos, K, Dunlop, D.C, Stuart, D.I, Barclay, A.N. | | Deposit date: | 2007-03-08 | | Release date: | 2007-03-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of the Macrophage Signal Regulatory Protein Alpha (Sirpalpha) Inhibitory Receptor Reveals a Binding Face Reminiscent of that Used by T Cell Receptors.
J.Biol.Chem., 282, 2007
|
|
8A5D
 
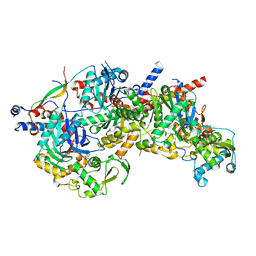 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of Chaetomium thermophilum INO80 | | Descriptor: | Actin, Actin related protein 4 (Arp4), Actin-related protein 8, ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-06-14 | | Release date: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
7MYN
 
 | | Cryo-EM Structure of p110alpha in complex with p85alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Yang, S, Hart, J.R, Xu, Y, Zou, X, Zhang, H, Zhou, Q, Xia, T, Zhang, Y, Yang, D, Wang, M.-W, Vogt, P.K. | | Deposit date: | 2021-05-21 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Cryo-EM structures of PI3K alpha reveal conformational changes during inhibition and activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MYO
 
 | | Cryo-EM structure of p110alpha in complex with p85alpha inhibited by BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Yang, S, Hart, J.R, Xu, Y, Zou, X, Zhang, H, Zhou, Q, Xia, T, Zhang, Y, Yang, D, Wang, M.-W, Vogt, P.K. | | Deposit date: | 2021-05-21 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structures of PI3K alpha reveal conformational changes during inhibition and activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2J7C
 
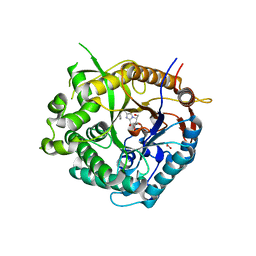 | | Beta-glucosidase from Thermotoga maritima in complex with phenylaminomethyl-derived glucoimidazole | | Descriptor: | (5R,6R,7S,8S)-3-(ANILINOMETHYL)-5,6,7,8-TETRAHYDRO-5-(HYDROXYMETHYL)-IMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, ACETATE ION, BETA-GLUCOSIDASE A, ... | | Authors: | Gloster, T.M, Zechel, D, Vasella, A, Davies, G.J. | | Deposit date: | 2006-10-06 | | Release date: | 2006-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Glycosidase Inhibition: An Assessment of the Binding of 18 Putative Transition-State Mimics.
J.Am.Chem.Soc., 129, 2007
|
|
8A6I
 
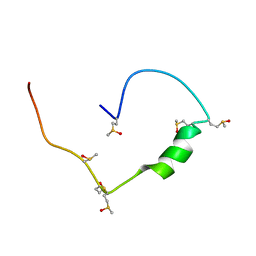 | | Structure of the low complexity domain of TDP-43 (fragment 309-350) with methionine sulfoxide modifications | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Carrasco, J, Anton, R, Pantoja-Uceda, D, Laurents, D.V, Oroz, J. | | Deposit date: | 2022-06-17 | | Release date: | 2023-02-08 | | Method: | SOLUTION NMR | | Cite: | Metamorphism in TDP-43 prion-like domain determines chaperone recognition.
Nat Commun, 14, 2023
|
|
2VH1
 
 | | Crystal structure of bacterial cell division protein FtsQ from E.coli | | Descriptor: | CELL DIVISION PROTEIN FTSQ | | Authors: | van den Ent, F, Vinkenvleugel, T, Ind, A, West, P, Veprintsev, D, Naninga, N, den Blaauwen, T, Lowe, J. | | Deposit date: | 2007-11-16 | | Release date: | 2008-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Mutational Analysis of Cell Division Protein Ftsq
Mol.Microbiol., 68, 2008
|
|
2VDU
 
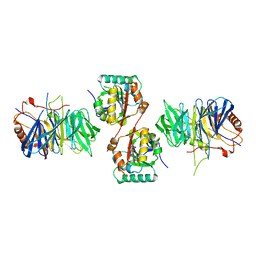 | | Structure of trm8-trm82, THE YEAST TRNA m7G methylation complex | | Descriptor: | PHOSPHATE ION, TRNA (GUANINE-N(7)-)-METHYLTRANSFERASE, TRNA (GUANINE-N(7)-)-METHYLTRANSFERASE-ASSOCIATED WD REPEAT PROTEIN TRM82 | | Authors: | Leulliot, N, Chaillet, M, Durand, D, Ulryck, N, Blondeau, K, Van Tilbeurgh, H. | | Deposit date: | 2007-10-11 | | Release date: | 2007-12-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Yeast tRNA M7G Methylation Complex.
Structure, 16, 2008
|
|
1NLD
 
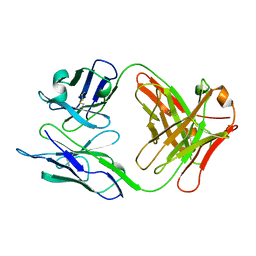 | | FAB FRAGMENT OF A NEUTRALIZING ANTIBODY DIRECTED AGAINST AN EPITOPE OF GP41 FROM HIV-1 | | Descriptor: | FAB1583 | | Authors: | Davies, C, Beauchamp, J.C, Emery, D, Rawas, A, Muirhead, H. | | Deposit date: | 1996-07-02 | | Release date: | 1996-12-23 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Fab fragment from a neutralizing monoclonal antibody directed against an epitope of gp41 from HIV-1.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
7NAV
 
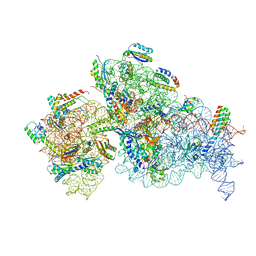 | | Bacterial 30S ribosomal subunit assembly complex state D (Consensus refinement) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7NAU
 
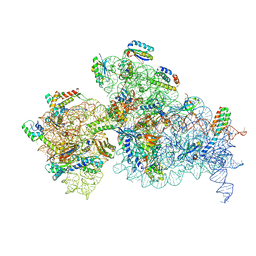 | | Bacterial 30S ribosomal subunit assembly complex state C (Consensus Refinement) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
