7O8I
 
 | | NmHR light state structure at 1 us after photoexcitation determined by serial femtosecond crystallography (with extrapolated, dark and light dataset) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8R
 
 | | NmHR light state structure at 27.5 ms (25 - 30 ms) after photoexcitation determined by serial millisecond crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8T
 
 | | NmHR light state structure at 37.5 ms (35 - 40 ms) after photoexcitation determined by serial millisecond crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8V
 
 | | NmHR light state structure at 55 ms (50 - 60 ms) after photoexcitation determined by serial millisecond crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8N
 
 | | NmHR light state structure at 7.5 ms (5 - 10 ms) after photoexcitation determined by serial millisecond crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8S
 
 | | NmHR light state structure at 32.5 ms (30 - 35 ms) after photoexcitation determined by serial millisecond crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8G
 
 | | NmHR light state structure at 10 ps after photoexcitation determined by serial femtosecond crystallography (with extrapolated, dark and light dataset) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8O
 
 | | NmHR light state structure at 12.5 ms (10 - 15 ms) after photoexcitation determined by serial millisecond crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8Y
 
 | | Anomalous bromide substructure of NmHR under dark state conditions determined at 13.7 keV with serial crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, BROMIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8U
 
 | | NmHR light state structure at 45 ms (40 - 50 ms) after photoexcitation determined by serial millisecond crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8J
 
 | | NmHR light state structure at 20 us after photoexcitation determined by serial femtosecond crystallography (with extrapolated, dark and light dataset) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8L
 
 | | NmHR dark state structure determined by serial millisecond crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
4YAY
 
 | | XFEL structure of human Angiotensin Receptor | | Descriptor: | 5,7-diethyl-1-{[2'-(1H-tetrazol-5-yl)biphenyl-4-yl]methyl}-3,4-dihydro-1,6-naphthyridin-2(1H)-one, Soluble cytochrome b562,Type-1 angiotensin II receptor | | Authors: | Zhang, H, Unal, H, Gati, C, Han, G.W, Zatsepin, N.A, James, D, Wang, D, Nelson, G, Weierstall, U, Messerschmidt, M, Williams, G.J, Boutet, S, Yefanov, O.M, White, T.A, Liu, W, Ishchenko, A, Tirupula, K.C, Desnoyer, R, Sawaya, M.C, Xu, Q, Coe, J, Cornrad, C.E, Fromme, P, Stevens, R.C, Katritch, V, Karnik, S.S, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2015-02-18 | | Release date: | 2015-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Angiotensin receptor revealed by serial femtosecond crystallography.
Cell, 161, 2015
|
|
6QLE
 
 | | Structure of inner kinetochore CCAN complex | | Descriptor: | Central kinetochore subunit CTF3,Inner kinetochore subunit CTF3,Central kinetochore subunit CTF3,Inner kinetochore subunit CTF3, Central kinetochore subunit MCM16,Central kinetochore subunit MCM16,Inner kinetochore subunit MCM16,Mcm16p, Inner kinetochore subunit AME1,Inner kinetochore subunit AME1,Inner kinetochore subunit AME1,Inner kinetochore subunit AME1, ... | | Authors: | Yan, K, Yang, J, Zhang, Z, McLaughlin, S.H, Chang, L, Fasci, D, Heck, A.J.R, Barford, D. | | Deposit date: | 2019-01-31 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome.
Nature, 574, 2019
|
|
5HKM
 
 | | DISCOVERY OF NOVEL 7-AZAINDOLES AS PDK1 INHIBITORS | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 4-ethyl-6-[5-(1H-pyrazol-4-yl)-1H-pyrrolo[2,3-b]pyridin-3-yl]pyrimidin-2-amine, SULFATE ION | | Authors: | Wucherer-Plietker, M, Esdar, C, Knoechel, T, Hillertz, P, Heinrich, T, Buchstaller, H.P, Greiner, H, Dorsch, D, Calderini, M, Bruge, D, Mueller, T.J.J, Graedler, U. | | Deposit date: | 2016-01-14 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of novel 7-azaindoles as PDK1 inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
4YMG
 
 | | Crystal structure of SAM-bound Podospora anserina methyltransferase PaMTH1 | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Putative SAM-dependent O-methyltranferase, ... | | Authors: | Kudlinzki, D, Linhard, V.L, Chatterjee, D, Saxena, K, Sreeramulu, S, Schwalbe, H. | | Deposit date: | 2015-03-06 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structure and Biophysical Characterization of the S-Adenosylmethionine-dependent O-Methyltransferase PaMTH1, a Putative Enzyme Accumulating during Senescence of Podospora anserina.
J.Biol.Chem., 290, 2015
|
|
5HO8
 
 | | DISCOVERY OF NOVEL 7-AZAINDOLES AS PDK1 INHIBITORS | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 4-butyl-6-(1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-2-amine, SULFATE ION | | Authors: | Wucherer-Plietker, M, Esdar, C, Knoechel, T, Hillertz, P, Heinrich, T, Buchstaller, H.P, Greiner, H, Dorsch, D, Calderini, M, Bruge, D, Mueller, T.J.J, Graedler, U. | | Deposit date: | 2016-01-19 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of novel 7-azaindoles as PDK1 inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
4YMH
 
 | | Crystal structure of SAH-bound Podospora anserina methyltransferase PaMTH1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Putative SAM-dependent O-methyltranferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Kudlinzki, D, Linhard, V.L, Chatterjee, D, Saxena, K, Sreeramulu, S, Schwalbe, H. | | Deposit date: | 2015-03-06 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.876 Å) | | Cite: | Structure and Biophysical Characterization of the S-Adenosylmethionine-dependent O-Methyltransferase PaMTH1, a Putative Enzyme Accumulating during Senescence of Podospora anserina.
J.Biol.Chem., 290, 2015
|
|
5BNJ
 
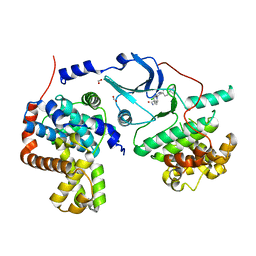 | | CDK8/CYCC IN COMPLEX WITH 8-{3-Chloro-5-[4-(1-methyl-1H-pyrazol-4-yl)-phenyl]-pyridin- 4-yl}-2,8-diaza-spiro[4.5]decan-1-one | | Descriptor: | 8-{3-chloro-5-[4-(1-methyl-1H-pyrazol-4-yl)phenyl]pyridin-4-yl}-2,8-diazaspiro[4.5]decan-1-one, Cyclin-C, Cyclin-dependent kinase 8, ... | | Authors: | Musil, D, Blagg, J, Wienke, D. | | Deposit date: | 2015-05-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | A selective chemical probe for exploring the role of CDK8 and CDK19 in human disease.
Nat.Chem.Biol., 11, 2015
|
|
6O0I
 
 | | NMR ensemble of computationally designed protein XAA | | Descriptor: | Design construct XAA | | Authors: | Wei, K.Y, Moschidi, D, Nerli, S, Sgourakis, N, Baker, D. | | Deposit date: | 2019-02-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Computational design of closely related proteins that adopt two well-defined but structurally divergent folds.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3BPX
 
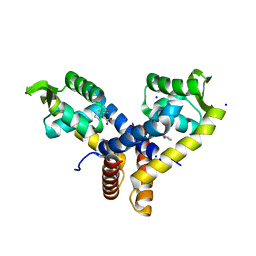 | | Crystal Structure of MarR | | Descriptor: | 2-HYDROXYBENZOIC ACID, SODIUM ION, Transcriptional regulator | | Authors: | Saridakis, V, Shahinas, D, Xu, X, Christendat, D. | | Deposit date: | 2007-12-19 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insight on the mechanism of regulation of the MarR family of proteins: high-resolution crystal structure of a transcriptional repressor from Methanobacterium thermoautotrophicum.
J.Mol.Biol., 377, 2008
|
|
8ONW
 
 | |
3AG9
 
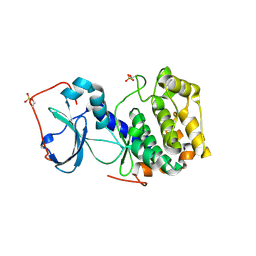 | | Complex of PKA with the bisubstrate protein kinase inhibitor ARC-1012 | | Descriptor: | (10R,20R,23R)-10-(4-aminobutyl)-1-[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-20,23-bis(3-carbamimidamidopropyl)-1,8,11,18,21-pentaoxo-2,9,12,19,22-pentaazatetracosan-24-amide, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Pflug, A, Lavogina, D, Uri, A, Engh, R.A, Bossemeyer, D. | | Deposit date: | 2010-03-26 | | Release date: | 2010-09-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diversity of bisubstrate binding modes of adenosine analogue-oligoarginine conjugates in protein kinase a and implications for protein substrate interactions.
J.Mol.Biol., 403, 2010
|
|
3BPV
 
 | | Crystal Structure of MarR | | Descriptor: | Transcriptional regulator | | Authors: | Saridakis, V, Shahinas, D, Xu, X, Christendat, D. | | Deposit date: | 2007-12-19 | | Release date: | 2008-05-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insight on the mechanism of regulation of the MarR family of proteins: high-resolution crystal structure of a transcriptional repressor from Methanobacterium thermoautotrophicum.
J.Mol.Biol., 377, 2008
|
|
3AQ1
 
 | | Open state monomer of a group II chaperonin from methanococcoides burtonii | | Descriptor: | Thermosome subunit | | Authors: | Harrop, S.J, Pilak, O, Siddiqui, K.S, De Francisci, D, Burg, D, Williams, T.J, Cavicchioli, R, Curmi, P.M. | | Deposit date: | 2010-10-24 | | Release date: | 2011-09-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.746 Å) | | Cite: | Chaperonins from an Antarctic archaeon are predominantly monomeric: crystal structure of an open state monomer.
ENVIRON.MICROBIOL., 13, 2011
|
|
