1B6J
 
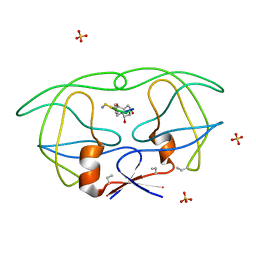 | | HIV-1 PROTEASE COMPLEXED WITH MACROCYCLIC PEPTIDOMIMETIC INHIBITOR 1 | | Descriptor: | CYCLIC PEPTIDE INHIBITOR, RETROPEPSIN, SULFATE ION | | Authors: | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | Deposit date: | 1999-01-17 | | Release date: | 2000-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease.
Biochemistry, 38, 1999
|
|
1R7C
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Minimized average structure, Sample in 50% tfe) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
1R7F
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Ensemble of 43 structures. Sample in 100mM SDS) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
8S5R
 
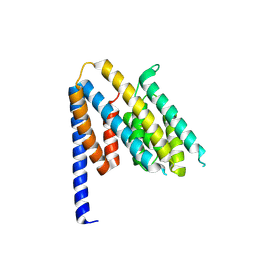 | | Structure of the Chlamydia pneumoniae effector SemD | | Descriptor: | Effector SemD | | Authors: | Kocher, F, Applegate, V, Reiners, J, Port, A, Spona, D, Haensch, S, Smits, S.H, Hegemann, J, Moelleken, K. | | Deposit date: | 2024-02-25 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Chlamydia pneumoniae effector SemD
To Be Published
|
|
5MW6
 
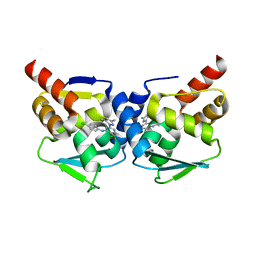 | | Crystal structure of the BCL6 BTB-domain with compound 1 | | Descriptor: | 5-chloranyl-~{N}-(4-chlorophenyl)-2-(3,5-dimethylpyrazol-1-yl)pyrimidin-4-amine, B-cell lymphoma 6 protein | | Authors: | Davies, D.R, Kessler, D. | | Deposit date: | 2017-01-18 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Chemically Induced Degradation of the Oncogenic Transcription Factor BCL6.
Cell Rep, 20, 2017
|
|
7P2N
 
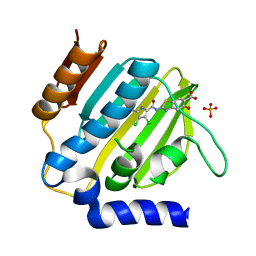 | | E.coli GyrB24 with inhibitor LSJ38 (EBL2684) | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-5-oxidanyl-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, PHOSPHATE ION | | Authors: | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Exploring the 5-Substituted 2-Aminobenzothiazole-Based DNA Gyrase B Inhibitors Active against ESKAPE Pathogens.
Acs Omega, 8, 2023
|
|
9CTX
 
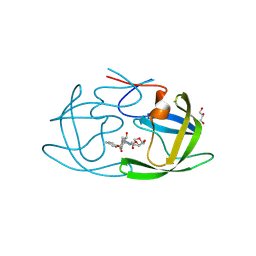 | | X-ray crystal structure of multi-drug resistant HIV-1 protease (P51) in complex with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, GLYCEROL, Protease | | Authors: | Hayashi, H, Yedidi, R, Bulut, H, Das, D, Mitsuya, H. | | Deposit date: | 2024-07-25 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Secondary amino-acid substitutions contribute to the emergence of HIV protease inhibitor resistance as directly as primary amino-acid substitutions.
To Be Published
|
|
1RNX
 
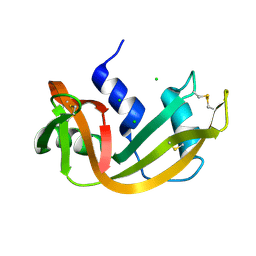 | | RIBONUCLEASE A CRYSTALLIZED FROM 3M SODIUM CHLORIDE, 30% AMMONIUM SULFATE | | Descriptor: | CHLORIDE ION, RIBONUCLEASE A | | Authors: | Fedorov, A.A, Joseph-Mccarthy, D, Fedorov, L, Sirakova, D, Graf, I, Almo, S.C. | | Deposit date: | 1996-04-23 | | Release date: | 1996-11-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ionic interactions in crystalline bovine pancreatic ribonuclease A.
Biochemistry, 35, 1996
|
|
8S5T
 
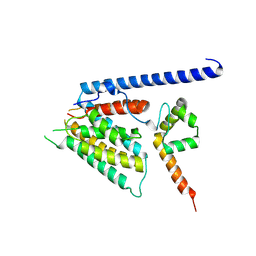 | | Structure of SemD in complex | | Descriptor: | Effector SemD, Neural Wiskott-Aldrich syndrome protein | | Authors: | Kocher, F, Applegate, V, Port, A, Reiners, J, Spona, D, Haensch, S, Smits, S.H, Hegemann, J, Moelleken, K. | | Deposit date: | 2024-02-25 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of SemD in complex
To Be Published
|
|
5VEW
 
 | | Structure of the human GLP-1 receptor complex with PF-06372222 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Glucagon-like peptide 1 receptor,Endolysin chimera, N-{4-[(R)-(3,3-dimethylcyclobutyl)({6-[4-(trifluoromethyl)-1H-imidazol-1-yl]pyridin-3-yl}amino)methyl]benzene-1-carbonyl}-beta-alanine, ... | | Authors: | Song, G, Yang, D, Wang, Y, Graaf, C.D, Zhou, Q, Jiang, S, Liu, K, Cai, X, Dai, A, Lin, G, Liu, D, Wu, F, Wu, Y, Zhao, S, Ye, L, Han, G.W, Lau, J, Wu, B, Hanson, M.A, Liu, Z.-J, Wang, M.-W, Stevens, R.C. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators.
Nature, 546, 2017
|
|
5O7N
 
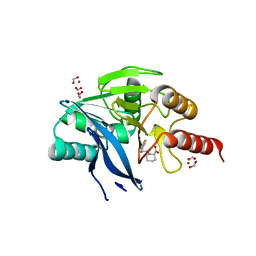 | | Beta-lactamase VIM-2 in complex with (2R)-1-(2-Benzyl-3-mercaptopropanoyl)piperidine-2-carboxylic acid | | Descriptor: | (2~{R})-1-[(2~{S})-2-(phenylmethyl)-3-sulfanyl-propanoyl]piperidine-2-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase VIM-2, ... | | Authors: | Buettner, D, Kramer, J.S, Pogoryelov, D, Proschak, E. | | Deposit date: | 2017-06-09 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Challenges in the Development of a Thiol-Based Broad-Spectrum Inhibitor for Metallo-beta-Lactamases.
Acs Infect Dis., 4, 2018
|
|
8E7O
 
 | | CRYSTAL STRUCTURE OF LYS48-LINKED TETRAUBIQUITIN | | Descriptor: | SULFATE ION, Ubiquitin | | Authors: | Lemma, B.E, Fushman, D. | | Deposit date: | 2022-08-24 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of selective recognition of Lys48-linked polyubiquitin by macrocyclic peptide inhibitors of proteasomal degradation.
Nat Commun, 14, 2023
|
|
1BH8
 
 | | HTAFII18/HTAFII28 HETERODIMER CRYSTAL STRUCTURE | | Descriptor: | TAFII18, TAFII28 | | Authors: | Birck, C, Poch, O, Romier, C, Ruff, M, Mengus, G, Lavigne, A.-C, Davidson, I, Moras, D. | | Deposit date: | 1998-06-16 | | Release date: | 1999-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human TAF(II)28 and TAF(II)18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family.
Cell(Cambridge,Mass.), 94, 1998
|
|
1BHE
 
 | |
1R4L
 
 | | Inhibitor Bound Human Angiotensin Converting Enzyme-Related Carboxypeptidase (ACE2) | | Descriptor: | (S,S)-2-{1-CARBOXY-2-[3-(3,5-DICHLORO-BENZYL)-3H-IMIDAZOL-4-YL]-ETHYLAMINO}-4-METHYL-PENTANOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Towler, P, Staker, B, Prasad, S.G, Menon, S, Ryan, D, Tang, J, Parsons, T, Fisher, M, Williams, D, Dales, N.A, Patane, M.A, Pantoliano, M.W. | | Deposit date: | 2003-10-07 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis.
J.Biol.Chem., 279, 2004
|
|
1R61
 
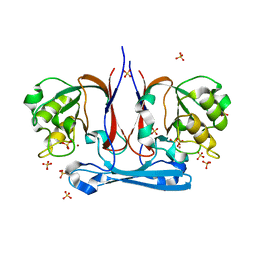 | | The structure of predicted metal-dependent hydrolase from Bacillus stearothermophilus | | Descriptor: | SULFATE ION, ZINC ION, metal-dependent hydrolase | | Authors: | Maderova, J, Borek, D, Tomchick, D, Joachimiak, A, Collart, F, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-10-14 | | Release date: | 2004-03-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of potential metal-dependent hydrolase with cyclase activity
To be Published
|
|
1AZ2
 
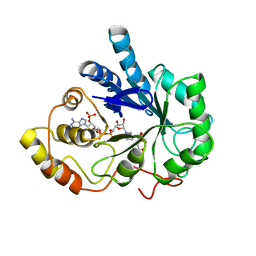 | | CITRATE BOUND, C298A/W219Y MUTANT HUMAN ALDOSE REDUCTASE | | Descriptor: | ALDOSE REDUCTASE, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H, Bohren, K.M, Ringe, D, Petsko, G.A, Gabbay, K.H. | | Deposit date: | 1997-11-24 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The alrestatin double-decker: binding of two inhibitor molecules to human aldose reductase reveals a new specificity determinant.
Biochemistry, 36, 1997
|
|
1B3B
 
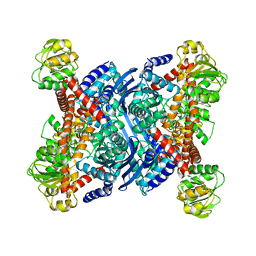 | | THERMOTOGA MARITIMA GLUTAMATE DEHYDROGENASE MUTANT N97D, G376K | | Descriptor: | PROTEIN (GLUTAMATE DEHYDROGENASE) | | Authors: | Knapp, S, Lebbink, J.H.G, Van Der Oost, J, Devos, W.M, Rice, D, Ladenstein, R. | | Deposit date: | 1998-12-07 | | Release date: | 1999-12-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Engineering activity and stability of Thermotoga maritima glutamate dehydrogenase. I. Introduction of a six-residue ion-pair network in the hinge region.
J.Mol.Biol., 280, 1998
|
|
1B6L
 
 | | HIV-1 PROTEASE COMPLEXED WITH MACROCYCLIC PEPTIDOMIMETIC INHIBITOR 4 | | Descriptor: | 1-[2-(8-CARBAMOYLMETHYL-6,9-DIOXO-2-OXA-7,10-DIAZA-BICYCLO[11.2.2]HEPTADECA- 1(16),13(17),14-TRIEN-11-YL)-2-HYDROXY-ETHYL]-PIPERIDINE-2-CARBOXYLIC ACID TERT-BUTYLAMIDE, RETROPEPSIN, SULFATE ION | | Authors: | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | Deposit date: | 1999-01-17 | | Release date: | 2000-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease.
Biochemistry, 38, 1999
|
|
1R7G
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Minimized average structure, Sample in 100mM DPC) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
1AUZ
 
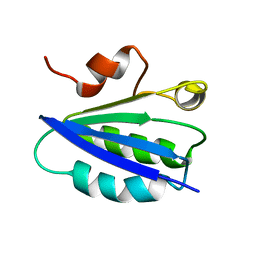 | | SOLUTION STRUCTURE OF SPOIIAA, A PHOSPHORYLATABLE COMPONENT OF THE SYSTEM THAT REGULATES TRANSCRIPTION FACTOR SIGMA-F OF BACILLUS SUBTILIS, NMR, 24 STRUCTURES | | Descriptor: | SPOIIAA | | Authors: | Kovacs, H, Comfort, D, Lord, M, Campbell, I.D, Yudkin, M.D. | | Deposit date: | 1997-09-08 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SpoIIAA, a phosphorylatable component of the system that regulates transcription factor sigmaF of Bacillus subtilis.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1AV3
 
 | | POTASSIUM CHANNEL BLOCKER KAPPA CONOTOXIN PVIIA FROM C. PURPURASCENS, NMR, 20 STRUCTURES | | Descriptor: | Kappa-conotoxin PVIIA | | Authors: | Scanlon, M.J, Naranjo, D, Thomas, L, Alewood, P.F, Lewis, R.J, Craik, D.J. | | Deposit date: | 1997-09-24 | | Release date: | 1998-10-14 | | Last modified: | 2020-12-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and proposed binding mechanism of a novel potassium channel toxin kappa-conotoxin PVIIA.
Structure, 5, 1997
|
|
1B26
 
 | | GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE | | Authors: | Knapp, S, Devos, W.M, Rice, D, Ladenstein, R. | | Deposit date: | 1998-12-04 | | Release date: | 1999-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of glutamate dehydrogenase from the hyperthermophilic eubacterium Thermotoga maritima at 3.0 A resolution.
J.Mol.Biol., 267, 1997
|
|
1B4K
 
 | | High resolution crystal structure of a MG2-dependent 5-aminolevulinic acid dehydratase | | Descriptor: | LAEVULINIC ACID, MAGNESIUM ION, PROTEIN (5-AMINOLEVULINIC ACID DEHYDRATASE), ... | | Authors: | Frankenberg, N, Jahn, D, Heinz, D.W. | | Deposit date: | 1998-12-22 | | Release date: | 1999-07-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | High resolution crystal structure of a Mg2+-dependent porphobilinogen synthase.
J.Mol.Biol., 289, 1999
|
|
1B5K
 
 | | 3,N4-ETHENO-2'-DEOXYCYTIDINE OPPOSITE THYMIDINE IN AN 11-MER DUPLEX, SOLUTION STRUCTURE FROM NMR AND MOLECULAR DYNAMICS | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*EDCP*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*TP*GP*TP*AP*CP*G)-3') | | Authors: | Cullinan, D, Korobka, A, Grollman, A.P, Patel, D.J, Eisenberg, M, De Santos, C.L. | | Deposit date: | 1999-01-07 | | Release date: | 1999-01-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of an oligodeoxynucleotide duplex containing the exocyclic lesion 3,N4-etheno-2'-deoxycytidine opposite thymidine: comparison with the duplex containing deoxyadenosine opposite the adduct.
Biochemistry, 35, 1996
|
|
