7LU0
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 4 | | 分子名称: | CALCIUM ION, NITRATE ION, Proteinase K | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-20 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.01 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LU1
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 5 | | 分子名称: | CALCIUM ION, NITRATE ION, Proteinase K | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-20 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.06 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LU2
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 6 | | 分子名称: | CALCIUM ION, NITRATE ION, Proteinase K | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-20 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.11 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LU3
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 7 | | 分子名称: | CALCIUM ION, NITRATE ION, Proteinase K | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-20 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
3RE7
 
 | | Copper (II) loaded Bullfrog Ferritin M chain | | 分子名称: | COPPER (II) ION, Ferritin, middle subunit | | 著者 | Bertini, I, Lalli, D, Mangani, S, Pozzi, C, Rosa, C, Turano, P. | | 登録日 | 2011-04-02 | | 公開日 | 2012-04-04 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.82 Å) | | 主引用文献 | Structural insights into the ferroxidase site of ferritins from higher eukaryotes.
J.Am.Chem.Soc., 134, 2012
|
|
3RJO
 
 | |
3RFN
 
 | | Epitope backbone grafting by computational design for improved presentation of linear epitopes on scaffold proteins | | 分子名称: | BB_1wnu_001, ZINC ION | | 著者 | Azoitei, M.L, Ban, Y.A, Julien, J.P, Bryson, S, Schroeter, A, Kalyuzhniy, O, Porter, J.R, Adachi, Y, Baker, D, Szabo, E, Pai, E.F, Schief, W.R. | | 登録日 | 2011-04-06 | | 公開日 | 2011-11-09 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope.
J.Mol.Biol., 415, 2012
|
|
7MBG
 
 | | SARS-CoV-2 Main protease in orthorhombic space group | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Douangamath, A, von Delft, F, Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Oliva, G, Godoy, A.S. | | 登録日 | 2021-03-31 | | 公開日 | 2021-04-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
3R0Y
 
 | | Crystal Structures of Multidrug-resistant HIV-1 Protease in Complex with Mechanism-Based Aspartyl Protease Inhibitors | | 分子名称: | Multidrug-resistant clinical isolate 769 HIV-1 Protease, N-[(2S)-1-{[(2S,3S)-3-hydroxy-5-oxo-5-{[(2R)-1-oxo-3-phenyl-1-(prop-2-yn-1-ylamino)propan-2-yl]amino}-1-phenylpentan-2-yl]amino}-3-methyl-1-oxobutan-2-yl]pyridine-2-carboxamide | | 著者 | Yedidi, R.S, Gupta, D, Liu, Z, Brunzelle, J, Kovari, I.A, Woster, P.M, Kovari, L.C. | | 登録日 | 2011-03-09 | | 公開日 | 2012-04-04 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structures of multidrug-resistant HIV-1 protease in complex with two potent anti-malarial compounds.
Biochem.Biophys.Res.Commun., 421, 2012
|
|
3R1M
 
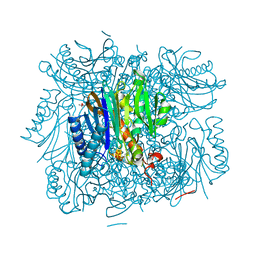 | | Structure of bifunctional fructose 1,6-bisphosphate aldolase/phosphatase (aldolase form) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,3-DIHYDROXYACETONEPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Fushinobu, S, Nishimasu, H, Hattori, D, Song, H.-J, Wakagi, T. | | 登録日 | 2011-03-10 | | 公開日 | 2011-10-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural basis for the bifunctionality of fructose-1,6-bisphosphate aldolase/phosphatase.
Nature, 478, 2011
|
|
3SJI
 
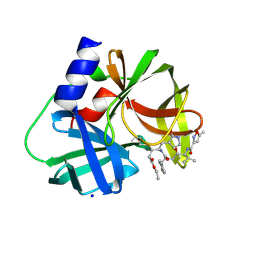 | | crystal structure of CVA16 3C in complex with Rupintrivir (AG7088) | | 分子名称: | 3C protease, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER, SODIUM ION | | 著者 | Lu, G, Qi, J, Chen, Z, Xu, X, Gao, F, Lin, D, Qian, W, Liu, H, Jiang, H, Yan, J, Gao, G.F. | | 登録日 | 2011-06-21 | | 公開日 | 2011-08-10 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.798 Å) | | 主引用文献 | Enterovirus 71 and Coxsackievirus A16 3C Proteases: Binding to Rupintrivir and Their Substrates and Anti-Hand, Foot, and Mouth Disease Virus Drug Design.
J.Virol., 85, 2011
|
|
3SMQ
 
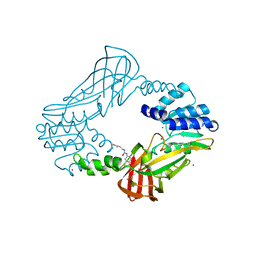 | | Crystal structure of protein arginine methyltransferase 3 | | 分子名称: | 1-(1,2,3-benzothiadiazol-6-yl)-3-[2-(cyclohex-1-en-1-yl)ethyl]urea, CHLORIDE ION, Protein arginine N-methyltransferase 3, ... | | 著者 | Dobrovetsky, E, Dong, A, Walker, J.R, Siarheyeva, A, Senisterra, G, Wasney, G.A, Smil, D, Bolshan, Y, Nguyen, K.T, Allali-Hassani, A, Hajian, T, Poda, G, Bountra, C, Weigelt, J, Edwards, A.M, Al-Awar, R, Brown, P.J, Schapira, M, Arrowsmith, C.H, Vedadi, M, Structural Genomics Consortium (SGC) | | 登録日 | 2011-06-28 | | 公開日 | 2011-08-31 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | An allosteric inhibitor of protein arginine methyltransferase 3.
Structure, 20, 2012
|
|
7K40
 
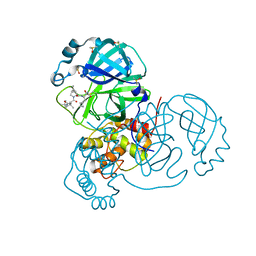 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor Boceprevir at 1.35 A Resolution | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, boceprevir (bound form) | | 著者 | Kumaran, D, Andi, B, Kreitler, D.F, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | 登録日 | 2020-09-14 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7LCC
 
 | | Helitron transposase bound to LTS | | 分子名称: | Helraiser K1068Q, LTS, ZINC ION | | 著者 | Kosek, D, Dyda, F. | | 登録日 | 2021-01-10 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.66 Å) | | 主引用文献 | The large bat Helitron DNA transposase forms a compact monomeric assembly that buries and protects its covalently bound 5'-transposon end.
Mol.Cell, 81, 2021
|
|
3SJ7
 
 | |
7LON
 
 | | Ornithine Aminotransferase (OAT) cocrystallized with its inactivator - (1S,3S)-3-amino-4-(difluoromethylene)cyclohexene-1-carboxylic acid | | 分子名称: | (1R,3S,4R)-3-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-4-methylcyclohexane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial, ... | | 著者 | Butrin, A, Zhu, W, Liu, D, Silverman, R. | | 登録日 | 2021-02-10 | | 公開日 | 2021-08-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Remarkable and Unexpected Mechanism for ( S )-3-Amino-4-(difluoromethylenyl)cyclohex-1-ene-1-carboxylic Acid as a Selective Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
7LOM
 
 | | Ornithine Aminotransferase (OAT) soaked with its inactivator - (1S,3S)-3-amino-4-(difluoromethylene)cyclohexene-1-carboxylic acid | | 分子名称: | (3~{S},4~{S})-4-methyl-3-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]cyclohexene-1-carboxylic acid, (4~{R})-4-(fluoranylmethyl)-3-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]cyclohexene-1-carboxylic acid, Ornithine aminotransferase, ... | | 著者 | Butrin, A, Zhu, W, Liu, D, Silverman, R. | | 登録日 | 2021-02-10 | | 公開日 | 2021-08-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Remarkable and Unexpected Mechanism for ( S )-3-Amino-4-(difluoromethylenyl)cyclohex-1-ene-1-carboxylic Acid as a Selective Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
3SEV
 
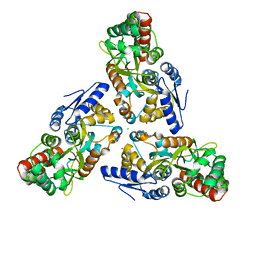 | | Zn-mediated Trimer of Maltose-binding Protein E310H/K314H by Synthetic Symmetrization | | 分子名称: | CHLORIDE ION, Maltose-binding periplasmic protein, ZINC ION, ... | | 著者 | Zhao, M, Soriaga, A.B, Laganowsky, A, Sawaya, M.R, Cascio, D, Yeates, T.O. | | 登録日 | 2011-06-11 | | 公開日 | 2011-09-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
3TBV
 
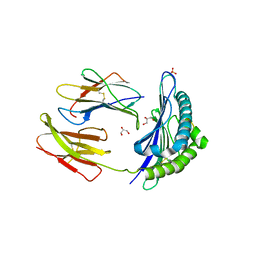 | | CRYSTAL STRUCTURE OF THE MURINE CLASS I MAJOR HISTOCOMPATIBILITY COMPLEX H-2DB IN COMPLEX WITH THE LCMV-DERIVED GP33 ALTERED PEPTIDE ligand (A2G,V3P,Y4A) | | 分子名称: | Beta-2-microglobulin, GLYCEROL, Glycoprotein G1, ... | | 著者 | Duru, A.D, Allerbring, E.B, Uchtenhagen, H, Mazumdar, P.A, Badia-Martinez, D, Madhurantakam, C, Sandalova, T, Nygren, P, Achour, A. | | 登録日 | 2011-08-08 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Conversion of a T cell viral antagonist into an agonist through higher stabilization and conserved molecular mimicry: Implications for TCR recognition
To be Published
|
|
3TGJ
 
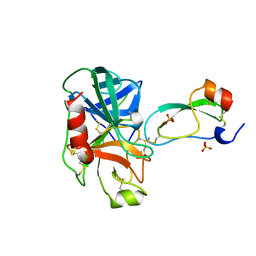 | | S195A TRYPSINOGEN COMPLEXED WITH BOVINE PANCREATIC TRYPSIN INHIBITOR (BPTI) | | 分子名称: | BOVINE PANCREATIC TRYPSIN INHIBITOR, CALCIUM ION, SULFATE ION, ... | | 著者 | Pasternak, A, Ringe, D, Hedstrom, L. | | 登録日 | 1998-07-16 | | 公開日 | 1998-12-23 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Comparison of Anionic and Cationic Trypsinogens: The Anionic Activation Domain is More Flexible in Solution and Differs in its Mode of Bpti Binding in the Crystal Structure
Protein Sci., 8, 1999
|
|
7LIV
 
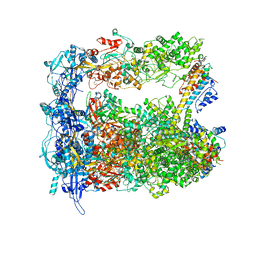 | | Structure of human transfer RNA visualized in the cytomegalovirus, a DNA virus | | 分子名称: | Major capsid protein, Small capsomere-interacting protein, Tegument protein pp150, ... | | 著者 | Liu, Y.T, Strugatsky, D, Liu, W, Zhou, Z.H. | | 登録日 | 2021-01-28 | | 公開日 | 2021-09-15 | | 最終更新日 | 2021-11-17 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structure of human cytomegalovirus virion reveals host tRNA binding to capsid-associated tegument protein pp150.
Nat Commun, 12, 2021
|
|
3T64
 
 | | 5'-Diphenyl Nucleoside Inhibitors of Plasmodium falciparum dUTPase | | 分子名称: | 2',5'-dideoxy-5'-[(diphenylmethyl)amino]uridine, 5'-(BENZHYDRYLAMINO)-2',5'-DIDEOXYURIDINE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | 著者 | Hampton, S.E, Baragana, B, Schipani, A, Bosch-Navarrete, C, Musso-Buendia, A, Recio, E, Kaiser, M, Whittingham, J.L, Roberts, S.M, Shevtsov, M, Brannigan, J.A, Kahnberg, P, Brun, R, Wilson, K.S, Gonzalez-Pacanowska, D, Johansson, N.G, Gilbert, I.H. | | 登録日 | 2011-07-28 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Design, synthesis, and evaluation of 5'-diphenyl nucleoside analogues as inhibitors of the Plasmodium falciparum dUTPase.
Chemmedchem, 6, 2011
|
|
3TH3
 
 | | Mg2+ Is Required for Optimal Folding of the Gamma-Carboxyglutamic Acid (Gla) Domains of Vitamin K-Dependent Clotting Factors At Physiological Ca2+ | | 分子名称: | CALCIUM ION, CHLORIDE ION, Coagulation factor VII heavy chain, ... | | 著者 | Vadivel, K, Agah, S, Cascio, D, Padmanabhan, K, Bajaj, S.P. | | 登録日 | 2011-08-18 | | 公開日 | 2012-08-22 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Mg2+ Is Required for Optimal Folding of the Gamma-Carboxyglutamic Acid (Gla)
Domains of Vitamin K-Dependent Clotting Factors At Physiological Ca2+
To be Published
|
|
7MHK
 
 | | Crystal Structure of Apo/Unliganded SARS-CoV-2 Main Protease (Mpro) at 310 K | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9601 Å) | | 主引用文献 | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHN
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 277 K | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1908 Å) | | 主引用文献 | The temperature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro )
Iucrj, 9, 2022
|
|
