8BTO
 
 | | Helical structure of BcThsA in complex with 1''-3'gcADPR | | 分子名称: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, NAD(+) hydrolase ThsA, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Tamulaitiene, G, Sasnauskas, G, Sabonis, D. | | 登録日 | 2022-11-29 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (2.96 Å) | | 主引用文献 | Activation of Thoeris antiviral system via SIR2 effector filament assembly.
Nature, 627, 2024
|
|
3R6T
 
 | | Rat catechol o-methyltransferase in complex with the bisubstrate inhibitor 4'-fluoro-4,5-dihydroxy-biphenyl-3-carboxylic acid {(E)-3-[(2S,4R,5R)-4-hydroxy-5-(6-methyl-purin-9-yl)-tetrahydro-furan-2-yl]-allyl}-amide | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 4'-fluoro-4,5-dihydroxy-N-{(2E)-3-[(2S,4R,5R)-4-hydroxy-5-(6-methyl-9H-purin-9-yl)tetrahydrofuran-2-yl]prop-2-en-1-yl}biphenyl-3-carboxamide, CHLORIDE ION, ... | | 著者 | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | 登録日 | 2011-03-22 | | 公開日 | 2012-02-01 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Catechol-O-methyltransferase in complex with substituted 3'-deoxyribose bisubstrate inhibitors.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2KL6
 
 | | Solution NMR structure of the CARDB domain of PF1109 from Pyrococcus furiosus. Northeast Structural Genomics Consortium target PfR193A | | 分子名称: | Uncharacterized protein | | 著者 | Aramini, J.M, Lee, D, Ciccosanti, C, Hamilton, K, Nair, R, Rost, B, Acton, T.B, Xiao, R, Swapna, G.V.T, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2009-06-30 | | 公開日 | 2009-08-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of the CARDB domain of PF1109 from Pyrococcus furiosus. Northeast Structural Genomics Consortium target PfR193A
To be Published
|
|
3DA1
 
 | | X-Ray structure of the glycerol-3-phosphate dehydrogenase from Bacillus halodurans complexed with FAD. Northeast Structural Genomics Consortium target BhR167. | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Glycerol-3-phosphate dehydrogenase | | 著者 | Kuzin, A.P, Abashidze, M, Seetharaman, J, Wang, D, Janjua, H, Owens, L, Xiao, R, Nair, R, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-05-28 | | 公開日 | 2008-07-29 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | X-Ray structure of the glycerol-3-phosphate dehydrogenase from Bacillus halodurans complexed with FAD. Northeast Structural Genomics Consortium target BhR167.
To be Published
|
|
3DIH
 
 | |
2K65
 
 | |
2K5N
 
 | | Solution NMR Structure of the N-Terminal Domain of Protein ECA1580 from Erwinia carotovora, Northeast Structural Genomics Consortium Target EwR156A | | 分子名称: | Putative cold-shock protein | | 著者 | Mills, J.L, Eletsky, A, Zhang, Q, Lee, D, Jiang, M, Ciccosanti, C, Xiao, R, Lui, J, Everett, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-06-30 | | 公開日 | 2008-08-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Structure of the Putative Cold Shock Protein from Erwinia carotovora: Northeast Structural Genomics Consortium Target EwR156a
To be Published
|
|
3DDB
 
 | |
5UMT
 
 | | Crystal structure of N-terminal domain of human FACT complex subunit SPT16 | | 分子名称: | 1,2-ETHANEDIOL, FACT complex subunit SPT16, GLYCEROL | | 著者 | Su, D, Hu, Q, Thompson, J.R, Botuyan, M.V, Mer, G. | | 登録日 | 2017-01-29 | | 公開日 | 2018-01-31 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.092 Å) | | 主引用文献 | Crystal structure of N-terminal domain of human FACT complex subunit SPT16
To Be Published
|
|
3DME
 
 | | Crystal structure of conserved exported protein from Bordetella pertussis. NorthEast Structural Genomics target BeR141 | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, L(+)-TARTARIC ACID, conserved exported protein | | 著者 | Seetharaman, J, Abashidze, M, Wang, D, Janjua, H, Foote, E.L, Xiao, R, Liu, J, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-06-30 | | 公開日 | 2008-08-26 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of conserved exported protein from Bordetella pertussis. NorthEast Structural Genomics target BeR141
To be Published
|
|
2K7H
 
 | | NMR solution structure of soybean allergen Gly m 4 | | 分子名称: | Stress-induced protein SAM22 | | 著者 | Berkner, H, Neudecker, P, Mittag, D, Ballmer-Weber, B.K, Schweimer, K, Vieths, S, Roesch, P. | | 登録日 | 2008-08-11 | | 公開日 | 2009-05-05 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Cross-reactivity of pollen and food allergens: soybean Gly m 4 is a member of the Bet v 1 superfamily and closely resembles yellow lupine proteins
Biosci.Rep., 29, 2009
|
|
3DF6
 
 | | The thermo- and acido-stable ORF-99 from the archaeal virus AFV1 | | 分子名称: | CALCIUM ION, ORF99 | | 著者 | Goulet, A, Spinelli, S, Prangishvili, D, van Tilbeurgh, H, Cambillau, C, Campanacci, V. | | 登録日 | 2008-06-11 | | 公開日 | 2009-06-16 | | 最終更新日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | The thermo- and acido-stable ORF-99 from the archaeal virus AFV1
Protein Sci., 18, 2009
|
|
3DFA
 
 | | Crystal structure of kinase domain of calcium-dependent protein kinase cgd3_920 from Cryptosporidium parvum | | 分子名称: | Calcium-dependent protein kinase cgd3_920 | | 著者 | Wernimont, A.K, Lew, J, Lin, Y.H, Hassanali, A, Khuu, C, Alam, Z, Kozieradzki, I, Cossar, D, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Wilkstrom, M, Edwards, A.M, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | 登録日 | 2008-06-11 | | 公開日 | 2008-07-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Crystal structure of kinase domain of calcium-dependent protein kinase cgd3_920 from Cryptosporidium parvum.
To be Published
|
|
8BH5
 
 | | SARS-CoV-2 BA.2.12.1 RBD in complex with Beta-27 Fab and C1 nanobody | | 分子名称: | Beta-27 heavy chain, Beta-27 light chain, GLYCEROL, ... | | 著者 | Huo, J, Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2022-10-29 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Humoral responses against SARS-CoV-2 Omicron BA.2.11, BA.2.12.1 and BA.2.13 from vaccine and BA.1 serum.
Cell Discov, 8, 2022
|
|
2KDF
 
 | | NMR structure of minor S5a (196-306):K48 linked diubiquitin species | | 分子名称: | 26S proteasome non-ATPase regulatory subunit 4, Ubiquitin | | 著者 | Zhang, N, Wang, Q, Ehlinger, A, Randles, L, Lary, J.W, Kang, Y, Haririnia, A, Cole, J.L, Fushman, D, Walters, K.J. | | 登録日 | 2009-01-06 | | 公開日 | 2009-09-01 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the s5a:k48-linked diubiquitin complex and its interactions with rpn13.
Mol.Cell, 35, 2009
|
|
3GCP
 
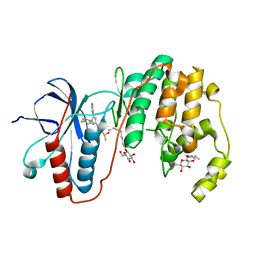 | | Human P38 MAP Kinase in Complex with SB203580 | | 分子名称: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | 著者 | Gruetter, C, Simard, J.R, Rauh, D. | | 登録日 | 2009-02-22 | | 公開日 | 2009-06-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Development of a fluorescent-tagged kinase assay system for the detection and characterization of allosteric kinase inhibitors.
J.Am.Chem.Soc., 131, 2009
|
|
3GDD
 
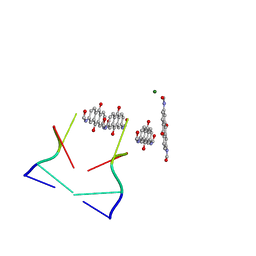 | | An inverted anthraquinone-DNA crystal structure | | 分子名称: | 5'-D(*(BRU)P*AP*GP*G)-3', MAGNESIUM ION, N,N'-(9,10-dioxo-9,10-dihydroanthracene-2,7-diyl)bis[2-(dimethylamino)acetamide] | | 著者 | Subirana, J.A, De Luchi, D, Wright, G, Gouyette, C. | | 登録日 | 2009-02-24 | | 公開日 | 2010-03-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of a stacked anthraquinone-DNA complex
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3R3C
 
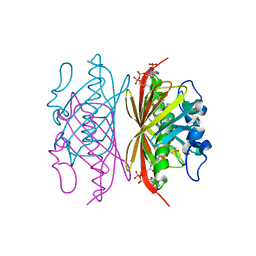 | | Crystal structure of Arthrobacter sp. strain SU 4-hydroxybenzoyl CoA thioesterase mutant H64A complexed with 4-hydroxyphenacyl CoA | | 分子名称: | 4-HYDROXYPHENACYL COENZYME A, 4-hydroxybenzoyl-CoA thioesterase | | 著者 | Holden, H.M, Thoden, J.B, Song, F, Zhuang, Z, Trujillo, M, Dunaway-Mariano, D. | | 登録日 | 2011-03-15 | | 公開日 | 2012-03-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The Catalytic Mechanism of the Hotdog-fold Enzyme Superfamily 4-Hydroxybenzoyl-CoA Thioesterase from Arthrobacter sp. Strain SU.
Biochemistry, 51, 2012
|
|
2KJD
 
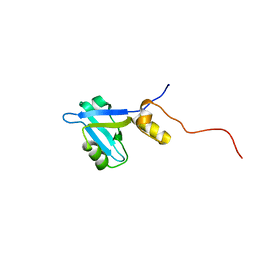 | |
3GGK
 
 | | Locating monovalent cations in one turn of G/C rich B-DNA | | 分子名称: | 5'-D(*CP*CP*AP*GP*GP*CP*CP*TP*GP*G) -3', MAGNESIUM ION, RUBIDIUM ION | | 著者 | Maehigashi, T, Moulaei, T, Watkins, D, Komeda, S, Williams, L.D. | | 登録日 | 2009-02-28 | | 公開日 | 2010-03-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (0.87 Å) | | 主引用文献 | Locating monovalent cations in one turn of G/C rich B-DNA
To be Published
|
|
8ADW
 
 | | Lipidic alpha-synuclein fibril - polymorph L1C | | 分子名称: | Alpha-synuclein | | 著者 | Frieg, B, Antonschmidt, L, Dienemann, C, Geraets, J.A, Najbauer, E.E, Matthes, D, de Groot, B.L, Andreas, L.B, Becker, S, Griesinger, C, Schroeder, G.F. | | 登録日 | 2022-07-11 | | 公開日 | 2022-10-12 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | The 3D structure of lipidic fibrils of alpha-synuclein.
Nat Commun, 13, 2022
|
|
2KMO
 
 | |
3UWY
 
 | |
5VHJ
 
 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | 分子名称: | 26S proteasome non-ATPase regulatory subunit 10, 26S proteasome non-ATPase regulatory subunit 2, 26S proteasome regulatory subunit 10B, ... | | 著者 | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | 登録日 | 2017-04-13 | | 公開日 | 2017-08-23 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (8.5 Å) | | 主引用文献 | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
8AB3
 
 | | Crystal Structure of the Lactate Dehydrogenase of Cyanobacterium Aponinum in complex with oxamate, NADH and FBP. | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1,6-di-O-phosphono-beta-D-fructofuranose, L-lactate dehydrogenase, ... | | 著者 | Robin, A.Y, Girard, E, Madern, D. | | 登録日 | 2022-07-04 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.616 Å) | | 主引用文献 | Deciphering Evolutionary Trajectories of Lactate Dehydrogenases Provides New Insights into Allostery.
Mol.Biol.Evol., 40, 2023
|
|
