1W8W
 
 | | CBM29-2 mutant Y46A: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | 分子名称: | NON-CATALYTIC PROTEIN 1 | | 著者 | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | 登録日 | 2004-09-30 | | 公開日 | 2005-03-22 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
7DZ9
 
 | | MbnABC complex | | 分子名称: | FE (III) ION, MbnA, MbnB, ... | | 著者 | Chao, D, Dan, Z, Yijun, G, Wei, C. | | 登録日 | 2021-01-25 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure and catalytic mechanism of the MbnBC holoenzyme required for methanobactin biosynthesis.
Cell Res., 32, 2022
|
|
1W2Y
 
 | | The crystal structure of a complex of Campylobacter jejuni dUTPase with substrate analogue dUpNHp | | 分子名称: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-DIPHOSPHATE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDE HYDROLASE, MAGNESIUM ION | | 著者 | Moroz, O.V, Harkiolaki, M, Galperin, M.Y, Vagin, A.A, Gonzalez-Pacanowska, D, Wilson, K.S. | | 登録日 | 2004-07-09 | | 公開日 | 2004-09-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | The Crystal Structure of a Complex of Campylobacter Jejuni Dutpase with Substrate Analogue Sheds Light on the Mechanism and Suggests the "Basic Module" for Dimeric D(C/U)Tpases
J.Mol.Biol., 342, 2004
|
|
1W5P
 
 | | Stepwise introduction of zinc binding site into porphobilinogen synthase of Pseudomonas aeruginosa (mutations A129C, D131C, D139C, P132E) | | 分子名称: | 1,2-ETHANEDIOL, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, FORMIC ACID, ... | | 著者 | Frere, F, Reents, H, Schubert, W.-D, Heinz, D.W, Jahn, D. | | 登録日 | 2004-08-09 | | 公開日 | 2005-01-19 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Tracking the Evolution of Porphobilinogen Synthase Metal Dependence in Vitro
J.Mol.Biol., 345, 2005
|
|
1W7A
 
 | | ATP bound MutS | | 分子名称: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP*TP* AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP*GP*GP*CP*AP*GP* CP*T)-3', ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Lamers, M.H, Georgijevic, D, Lebbink, J, Winterwerp, H.H.K, Agianian, B, de Wind, N, Sixma, T.K. | | 登録日 | 2004-08-31 | | 公開日 | 2004-09-10 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | ATP Increases the Affinity between Muts ATPase Domains: Implications for ATP Hydrolysis and Conformational Changes
J.Biol.Chem., 279, 2004
|
|
1W90
 
 | | CBM29-2 mutant D114A: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | 分子名称: | 1,2-ETHANEDIOL, NON-CATALYTIC PROTEIN 1, SODIUM ION | | 著者 | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | 登録日 | 2004-10-01 | | 公開日 | 2005-03-18 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1W8Z
 
 | | CBM29-2 mutant K85A: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | 分子名称: | NON CATALYTIC PROTEIN 1 | | 著者 | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | 登録日 | 2004-10-01 | | 公開日 | 2005-03-22 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
6SX2
 
 | | dsRNA recognition by R38AK41A mutant of H7N1 NS1 RNA Binding Domain | | 分子名称: | 1,2-ETHANEDIOL, Non-structural protein 1, RNA (5'-R(*GP*GP*UP*AP*AP*CP*UP*GP*UP*UP*AP*CP*AP*GP*UP*UP*AP*CP*C)-3') | | 著者 | Coste, F, Wacquiez, A, Marc, D, Castaing, B. | | 登録日 | 2019-09-24 | | 公開日 | 2020-10-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure and Sequence Determinants Governing the Interactions of RNAs with Influenza A Virus Non-Structural Protein NS1.
Viruses, 12, 2020
|
|
1W9F
 
 | | CBM29-2 mutant R112A: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | 分子名称: | NON CATALYTIC PROTEIN 1 | | 著者 | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | 登録日 | 2004-10-12 | | 公開日 | 2005-03-22 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1WBB
 
 | | Crystal structure of E. coli DNA mismatch repair enzyme MutS, E38A mutant, in complex with a G.T mismatch | | 分子名称: | 5'-D(*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP *GP*GP*CP*AP*GP*CP*T)-3', 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Natrajan, G, Georgijevic, D, Lebbink, J.H.G, Winterwerp, H.H.K, de Wind, N, Sixma, T.K. | | 登録日 | 2004-10-31 | | 公開日 | 2006-01-18 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Dual Role of Muts Glutamate 38 in DNA Mismatch Discrimination and in the Authorization of Repair.
Embo J., 25, 2006
|
|
1WBD
 
 | | Crystal structure of E. coli DNA mismatch repair enzyme MutS, E38Q mutant, in complex with a G.T mismatch | | 分子名称: | 5'-D(*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP *GP*GP*CP*AP*GP*CP*T)-3', 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Natrajan, G, Georgijevic, D, Lebbink, J.H.G, Winterwerp, H.H.K, de Wind, N, Sixma, T.K. | | 登録日 | 2004-10-31 | | 公開日 | 2006-01-18 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Dual Role of Muts Glutamate 38 in DNA Mismatch Discrimination and in the Authorization of Repair.
Embo J., 25, 2006
|
|
1W4W
 
 | | Ferric horseradish peroxidase C1A in complex with formate | | 分子名称: | CALCIUM ION, FORMIC ACID, HORSERADISH PEROXIDASE C1A, ... | | 著者 | Carlsson, G.H, Nicholls, P, Svistunenko, D, Berglund, G.I, Hajdu, J. | | 登録日 | 2004-08-03 | | 公開日 | 2005-01-19 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Complexes of Horseradish Peroxidase with Formate, Acetate, and Carbon Monoxide
Biochemistry, 44, 2005
|
|
1W54
 
 | | Stepwise introduction of a zinc binding site into Porphobilinogen synthase from Pseudomonas aeruginosa (mutation D139C) | | 分子名称: | DELTA-AMINOLEVULINIC ACID DEHYDRATASE, FORMIC ACID, MAGNESIUM ION, ... | | 著者 | Frere, F, Reents, H, Schubert, W.-D, Heinz, D.W, Jahn, D. | | 登録日 | 2004-08-05 | | 公開日 | 2005-01-19 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Tracking the Evolution of Porphobilinogen Synthase Metal Dependence in Vitro
J.Mol.Biol., 345, 2005
|
|
3WIS
 
 | | Crystal structure of Burkholderia xenovorans DmrB in complex with FMN: A Cubic Protein Cage for Redox Transfer | | 分子名称: | FLAVIN MONONUCLEOTIDE, Putative dihydromethanopterin reductase (AfpA), SULFATE ION | | 著者 | Bobik, T.A, Cascio, D, Jorda, J, McNamara, D.E, Bustos, C, Wang, T.C, Rasche, M.E, Yeates, T.O. | | 登録日 | 2013-09-25 | | 公開日 | 2014-02-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Structure of dihydromethanopterin reductase, a cubic protein cage for redox transfer
J.Biol.Chem., 289, 2014
|
|
1W9H
 
 | | The Structure of a Piwi protein from Archaeoglobus fulgidus. | | 分子名称: | CADMIUM ION, CHLORIDE ION, HYPOTHETICAL PROTEIN AF1318, ... | | 著者 | Parker, J.S, Roe, S.M, Barford, D. | | 登録日 | 2004-10-13 | | 公開日 | 2005-01-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal Structure of a Piwi Protein Suggests Mechanisms for Sirna Recognition and Slicer Activity
Embo J., 23, 2004
|
|
1W1H
 
 | | Crystal Structure of the PDK1 Pleckstrin Homology (PH) domain | | 分子名称: | 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE-1, GLYCEROL, SULFATE ION | | 著者 | Komander, D, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | 登録日 | 2004-06-21 | | 公開日 | 2004-11-19 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structural Insights Into the Regulation of Pdk1 by Phosphoinositides and Inositol Phosphates
Embo J., 23, 2004
|
|
4L3B
 
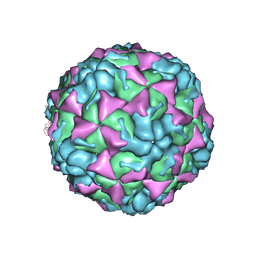 | | X-ray structure of the HRV2 A particle uncoating intermediate | | 分子名称: | Protein VP1, Protein VP2, Protein VP3 | | 著者 | Vives-Adrian, L, Querol-Audi, J, Garriga, D, Pous, J, Verdaguer, N. | | 登録日 | 2013-06-05 | | 公開日 | 2013-11-27 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (6.5 Å) | | 主引用文献 | Uncoating of common cold virus is preceded by RNA switching as determined by X-ray and cryo-EM analyses of the subviral A-particle.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1YSD
 
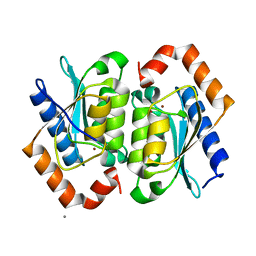 | | Yeast Cytosine Deaminase Double Mutant | | 分子名称: | CALCIUM ION, Cytosine deaminase, ZINC ION | | 著者 | Korkegian, A, Black, M.E, Baker, D, Stoddard, B.L. | | 登録日 | 2005-02-08 | | 公開日 | 2005-05-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Computational thermostabilization of an enzyme.
Science, 308, 2005
|
|
1YSP
 
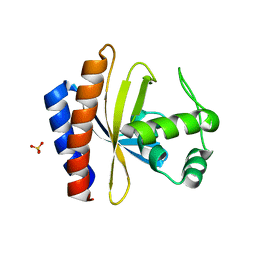 | | Crystal structure of the C-terminal domain of E. coli transcriptional regulator KdgR. | | 分子名称: | SULFATE ION, Transcriptional regulator kdgR | | 著者 | Bochkarev, A, Lunin, V.V, Ezersky, A, Evdokimova, E, Skarina, T, Xu, X, Borek, D, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2005-02-08 | | 公開日 | 2005-03-22 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural study of effector binding specificity in IclR transcriptional regulators
To be Published
|
|
2O44
 
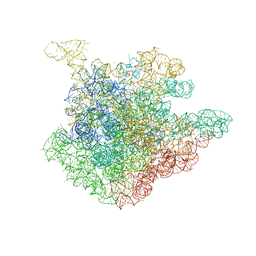 | | Structure of 23S rRNA of the large ribosomal subunit from Deinococcus radiodurans in complex with the macrolide josamycin | | 分子名称: | (2S,3S,4R,6S)-6-{[(2R,3S,4R,5R,6S)-6-{[(4R,5S,6S,7R,9R,10S,12E,14Z,16R)-4-(ACETYLOXY)-10-HYDROXY-5-METHOXY-9,16-DIMETHYL-2-OXO-7-(2-OXOETHYL)OXACYCLOHEXADECA-12,14-DIEN-6-YL]OXY}-4-(DIMETHYLAMINO)-5-HYDROXY-2-METHYLTETRAHYDRO-2H-PYRAN-3-YL]OXY}-4-HYDROXY-2,4-DIMETHYLTETRAHYDRO-2H-PYRAN-3-YL 3-METHYLBUTANOATE, 23S rRNA | | 著者 | Pyetan, E, Daram, D, Auerbach-Nevo, T, Yonath, A. | | 登録日 | 2006-12-03 | | 公開日 | 2007-12-04 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Chemical parameters influencing fine tuning in the binding of Macrolide antibiotics to the ribosomal tunnel
TO BE PUBLISHED
|
|
1YT8
 
 | |
1YNP
 
 | | aldo-keto reductase AKR11C1 from Bacillus halodurans (apo form) | | 分子名称: | GLYCEROL, SODIUM ION, SULFATE ION, ... | | 著者 | Marquardt, T, Kostrewa, D, Winkler, F.K, Li, X.D. | | 登録日 | 2005-01-25 | | 公開日 | 2005-12-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | High-resolution Crystal Structure of AKR11C1 from Bacillus halodurans: An NADPH-dependent 4-Hydroxy-2,3-trans-nonenal Reductase
J.Mol.Biol., 354, 2005
|
|
1YO4
 
 | |
1YT4
 
 | | Crystal structure of TEM-76 beta-lactamase at 1.4 Angstrom resolution | | 分子名称: | Beta-lactamase TEM | | 著者 | Thomas, V.L, Golemi-Kotra, D, Kim, C, Vakulenko, S.B, Mobashery, S, Shoichet, B.K. | | 登録日 | 2005-02-09 | | 公開日 | 2005-07-12 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural Consequences of the Inhibitor-Resistant Ser130Gly Substitution in TEM beta-Lactamase.
Biochemistry, 44, 2005
|
|
2ODA
 
 | | Crystal Structure of PSPTO_2114 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Hypothetical protein PSPTO_2114, MAGNESIUM ION | | 著者 | Peisach, E, Allen, K.N, Dunaway-Mariano, D, Wang, L, Burroughs, A.M, Aravind, L. | | 登録日 | 2006-12-22 | | 公開日 | 2007-09-04 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The X-ray crystallographic structure and activity analysis of a Pseudomonas-specific subfamily of the HAD enzyme superfamily evidences a novel biochemical function.
Proteins, 70, 2007
|
|
