7ZPI
 
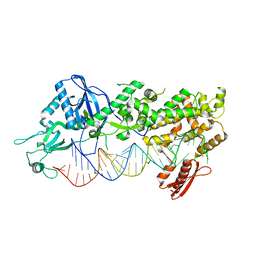 | | Mammalian Dicer in the "dicing state" with pre-miR-15a substrate | | 分子名称: | 59-nt precursor of miR-15a, Endoribonuclease Dicer | | 著者 | Zanova, M, Zapletal, D, Kubicek, K, Stefl, R, Pinkas, M, Novacek, J. | | 登録日 | 2022-04-27 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (5.91 Å) | | 主引用文献 | Structural and functional basis of mammalian microRNA biogenesis by Dicer.
Mol.Cell, 82, 2022
|
|
5ZC2
 
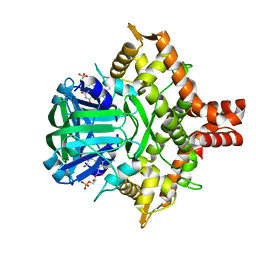 | | Acinetobacter baumannii p-hydroxyphenylacetate 3-hydroxylase (HPAH), reductase component (C1) | | 分子名称: | FLAVIN MONONUCLEOTIDE, p-hydroxyphenylacetate 3-hydroxylase, reductase component | | 著者 | Yuenyao, A, Petchyam, N, Chaiyen, P, Pakotiprapha, D. | | 登録日 | 2018-02-14 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.898 Å) | | 主引用文献 | Crystal structure of the flavin reductase of Acinetobacter baumannii p-hydroxyphenylacetate 3-hydroxylase (HPAH) and identification of amino acid residues underlying its regulation by aromatic ligands
Arch. Biochem. Biophys., 653, 2018
|
|
1A8G
 
 | | HIV-1 PROTEASE IN COMPLEX WITH SDZ283-910 | | 分子名称: | HIV-1 PROTEASE, benzyl [(1R)-1-({(1S,2S,3S)-1-benzyl-2-hydroxy-4-({(1S)-1-[(2-hydroxy-4-methoxybenzyl)carbamoyl]-2-methylpropyl}amino)-3-[(4-methoxybenzyl)amino]-4-oxobutyl}carbamoyl)-2,2-dimethylpropyl]carbamate | | 著者 | Kallen, J, Billich, A, Scholz, D, Auer, M, Kungl, A. | | 登録日 | 1998-03-24 | | 公開日 | 1998-07-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | X-ray structure and conformational dynamics of the HIV-1 protease in complex with the inhibitor SDZ283-910: agreement of time-resolved spectroscopy and molecular dynamics simulations.
J.Mol.Biol., 286, 1999
|
|
3KU4
 
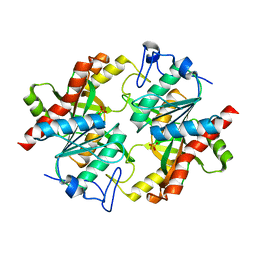 | | Trapping of an oxocarbenium ion intermediate in UP crystals | | 分子名称: | SULFATE ION, Uridine phosphorylase | | 著者 | Paul, D, O'Leary, S, Rajashankar, K, Bu, W, Toms, A, Settembre, E, Sanders, J, Begley, T.P, Ealick, S.E. | | 登録日 | 2009-11-26 | | 公開日 | 2010-04-28 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.099 Å) | | 主引用文献 | Glycal formation in crystals of uridine phosphorylase.
Biochemistry, 49, 2010
|
|
8CCI
 
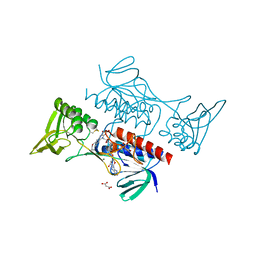 | | Crystal structure of Mycobacterium smegmatis thioredoxin reductase | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Fuesser, F.T, Koch, O, Kuemmel, D. | | 登録日 | 2023-01-27 | | 公開日 | 2023-07-26 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Novel starting points for fragment-based drug design against mycobacterial thioredoxin reductase identified using crystallographic fragment screening.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
1AHP
 
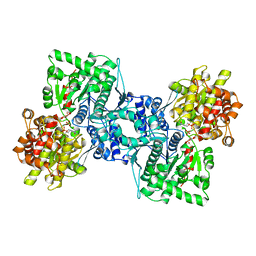 | | OLIGOSACCHARIDE SUBSTRATE BINDING IN ESCHERICHIA COLI MALTODEXTRIN PHSPHORYLASE | | 分子名称: | E.COLI MALTODEXTRIN PHOSPHORYLASE, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | 著者 | O'Reilly, M, Watson, K.A, Schinzel, R, Palm, D, Johnson, L.N. | | 登録日 | 1997-04-10 | | 公開日 | 1997-10-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Oligosaccharide substrate binding in Escherichia coli maltodextrin phosphorylase.
Nat.Struct.Biol., 4, 1997
|
|
5X5F
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 2 | | 分子名称: | S protein | | 著者 | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | 登録日 | 2017-02-15 | | 公開日 | 2017-05-03 | | 最終更新日 | 2017-05-24 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
6GYU
 
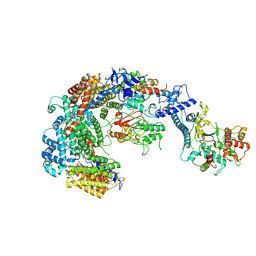 | | Cryo-EM structure of the CBF3-msk complex of the budding yeast kinetochore | | 分子名称: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | 著者 | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | 登録日 | 2018-07-02 | | 公開日 | 2018-12-05 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
1A6E
 
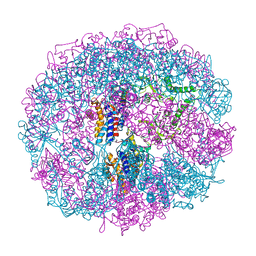 | | THERMOSOME-MG-ADP-ALF3 COMPLEX | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | 著者 | Ditzel, L, Loewe, J, Stock, D, Stetter, K.-O, Huber, H, Huber, R, Steinbacher, S. | | 登録日 | 1998-02-24 | | 公開日 | 1999-03-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT.
Cell(Cambridge,Mass.), 93, 1998
|
|
1A6D
 
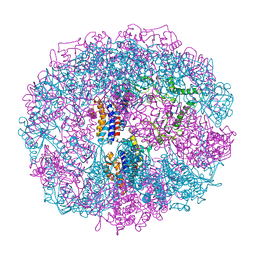 | | THERMOSOME FROM T. ACIDOPHILUM | | 分子名称: | THERMOSOME (ALPHA SUBUNIT), THERMOSOME (BETA SUBUNIT) | | 著者 | Ditzel, L, Loewe, J, Stock, D, Stetter, K.-O, Huber, H, Huber, R, Steinbacher, S. | | 登録日 | 1998-02-24 | | 公開日 | 1999-03-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT.
Cell(Cambridge,Mass.), 93, 1998
|
|
7ZDZ
 
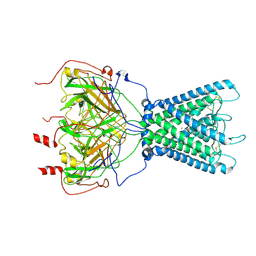 | | Cryo-EM structure of the human inward-rectifier potassium 2.1 channel (Kir2.1) | | 分子名称: | Inward rectifier potassium channel 2, POTASSIUM ION, STRONTIUM ION | | 著者 | Fernandes, C.A.H, Venien-Bryan, C, Fagnen, C, Zuniga, D. | | 登録日 | 2022-03-30 | | 公開日 | 2022-09-28 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Cryo-electron microscopy unveils unique structural features of the human Kir2.1 channel.
Sci Adv, 8, 2022
|
|
1AIK
 
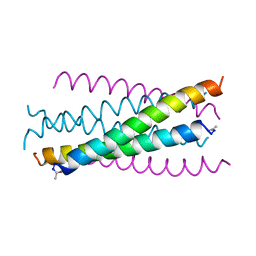 | | HIV GP41 CORE STRUCTURE | | 分子名称: | HIV-1 GP41 GLYCOPROTEIN | | 著者 | Chan, D.C, Fass, D, Berger, J.M, Kim, P.S. | | 登録日 | 1997-04-20 | | 公開日 | 1997-06-16 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Core structure of gp41 from the HIV envelope glycoprotein.
Cell(Cambridge,Mass.), 89, 1997
|
|
1A0E
 
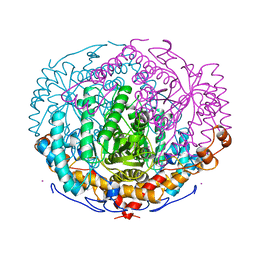 | | XYLOSE ISOMERASE FROM THERMOTOGA NEAPOLITANA | | 分子名称: | COBALT (II) ION, XYLOSE ISOMERASE | | 著者 | Gallay, O, Chopra, R, Conti, E, Brick, P, Blow, D. | | 登録日 | 1997-11-28 | | 公開日 | 1998-06-03 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal Structures of Class II Xylose Isomerases from Two Thermophiles and a Hyperthermophile
To be Published
|
|
1A8H
 
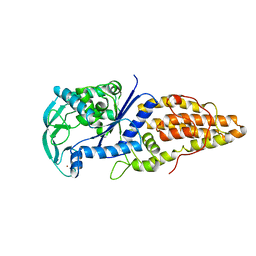 | | METHIONYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS | | 分子名称: | METHIONYL-TRNA SYNTHETASE, ZINC ION | | 著者 | Sugiura, I, Nureki, O, Ugaji, Y, Kuwabara, S, Lober, B, Giege, R, Moras, D, Yokoyama, S, Konno, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 1998-03-26 | | 公開日 | 1999-05-04 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The 2.0 A crystal structure of Thermus thermophilus methionyl-tRNA synthetase reveals two RNA-binding modules.
Structure, 8, 2000
|
|
5XJ6
 
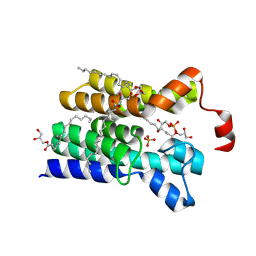 | | Crystal structure of PlsY (YgiH), an integral membrane glycerol 3-phosphate acyltransferase - the glycerol 3-phosphate form | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Glycerol-3-phosphate acyltransferase, PHOSPHATE ION, ... | | 著者 | Li, Z, Tang, Y, Li, D. | | 登録日 | 2017-04-30 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Structural insights into the committed step of bacterial phospholipid biosynthesis.
Nat Commun, 8, 2017
|
|
1A3D
 
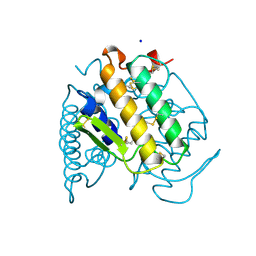 | | PHOSPHOLIPASE A2 (PLA2) FROM NAJA NAJA VENOM | | 分子名称: | PHOSPHOLIPASE A2, SODIUM ION | | 著者 | Segelke, B.W, Nguyen, D, Chee, R, Xuong, H.N, Dennis, E.A. | | 登録日 | 1998-01-20 | | 公開日 | 1998-04-29 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structures of two novel crystal forms of Naja naja naja phospholipase A2 lacking Ca2+ reveal trimeric packing.
J.Mol.Biol., 279, 1998
|
|
5XI2
 
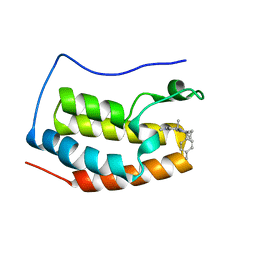 | | BRD4 bound with compound Bdi2 | | 分子名称: | (3~{R})-4-cyclopropyl-1,3-dimethyl-6-[[(1~{S})-1-(4-methylphenyl)ethyl]amino]-3~{H}-quinoxalin-2-one, Bromodomain-containing protein 4 | | 著者 | Xiong, B, Cao, D, Li, Y. | | 登録日 | 2017-04-25 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.909 Å) | | 主引用文献 | BRD4 bound with compound Bdi2
To Be Published
|
|
5XJ8
 
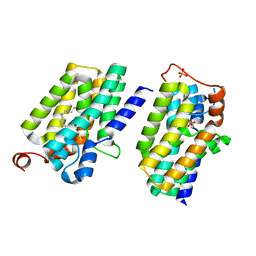 | | Crystal structure of PlsY (YgiH), an integral membrane glycerol 3-phosphate acyltransferase - the lysphosphatidic acid form | | 分子名称: | (2R)-2-hydroxy-3-(phosphonooxy)propyl hexadecanoate, Glycerol-3-phosphate acyltransferase, PHOSPHATE ION | | 著者 | Li, Z, Tang, Y, Li, D. | | 登録日 | 2017-04-30 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Structural insights into the committed step of bacterial phospholipid biosynthesis.
Nat Commun, 8, 2017
|
|
5CM5
 
 | | Structure of Hydroxyethylthiazole Kinase ThiM from Staphylococcus aureus | | 分子名称: | Hydroxyethylthiazole kinase | | 著者 | Drebes, J, Kuenz, M, Eberle, R.J, Oberthuer, D, Cang, H, Wrenger, C, Betzel, C. | | 登録日 | 2015-07-16 | | 公開日 | 2016-03-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Structure of ThiM from Vitamin B1 biosynthetic pathway of Staphylococcus aureus - Insights into a novel pro-drug approach addressing MRSA infections.
Sci Rep, 6, 2016
|
|
7JVC
 
 | | SARS-CoV-2 spike in complex with the S2A4 neutralizing antibody Fab fragment | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S2A4 Fab heavy chain, ... | | 著者 | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | 登録日 | 2020-08-20 | | 公開日 | 2020-10-14 | | 最終更新日 | 2021-06-23 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7BJP
 
 | | The cryo-EM structure of vesivirus 2117, an adventitious agent and possible cause of haemorrhagic gastroenteritis in dogs. | | 分子名称: | Capsid protein | | 著者 | Sutherland, H, Conley, M.J, Emmott, E, Streetley, J, Goodfellow, I.G, Bhella, D. | | 登録日 | 2021-01-14 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.65 Å) | | 主引用文献 | The Cryo-EM Structure of Vesivirus 2117 Highlights Functional Variations in Entry Pathways for Viruses in Different Clades of the Vesivirus Genus.
J.Virol., 95, 2021
|
|
7BXW
 
 | |
1CAQ
 
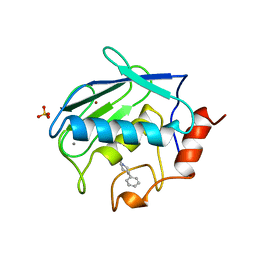 | | X-RAY STRUCTURE OF HUMAN STROMELYSIN CATALYTIC DOMAIN COMPLEXES WITH NON-PEPTIDE INHIBITORS: IMPLICATION FOR INHIBITOR SELECTIVITY | | 分子名称: | 3-(1H-INDOL-3-YL)-2-[4-(4-PHENYL-PIPERIDIN-1-YL)-BENZENESULFONYLAMINO]-PROPIONIC ACID, CALCIUM ION, PROTEIN (STROMELYSIN-1), ... | | 著者 | Pavlovsky, A.G, Williams, M.G, Ye, Q.-Z, Ortwine, D.F, Purchase II, C.F, White, A.D, Dhanaraj, V, Roth, B.D, Johnson, L.L, Hupe, D, Humblet, C, Blundell, T.L. | | 登録日 | 1999-02-23 | | 公開日 | 1999-07-07 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | X-ray structure of human stromelysin catalytic domain complexed with nonpeptide inhibitors: implications for inhibitor selectivity.
Protein Sci., 8, 1999
|
|
7JV6
 
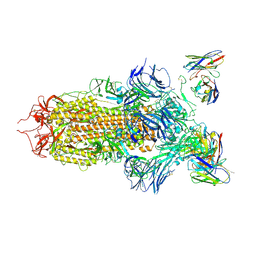 | | SARS-CoV-2 spike in complex with the S2H13 neutralizing antibody (closed conformation) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S2H13 Fab heavy chain, ... | | 著者 | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | 登録日 | 2020-08-20 | | 公開日 | 2020-10-14 | | 最終更新日 | 2021-06-23 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
5XRR
 
 | | Crystal structure of FUS (54-59) SYSSYG | | 分子名称: | RNA-binding protein FUS, ZINC ION | | 著者 | Zhao, M, Gui, X, Li, D, Liu, C. | | 登録日 | 2017-06-09 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.503 Å) | | 主引用文献 | Atomic structures of FUS LC domain segments reveal bases for reversible amyloid fibril formation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
