4ZIB
 
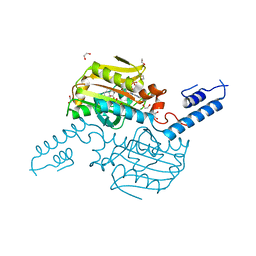 | | Crystal Structure of the C-terminal domain of PylRS mutant bound with 3-benzothienyl-l-alanine and ATP | | 分子名称: | 1,2-ETHANEDIOL, 3-(1-benzothiophen-3-yl)-L-alanine, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Nakamura, A, Guo, L.T, Wang, Y.S, Soll, D. | | 登録日 | 2015-04-28 | | 公開日 | 2016-03-02 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.054 Å) | | 主引用文献 | Probing the active site tryptophan of Staphylococcus aureus thioredoxin with an analog.
Nucleic Acids Res., 43, 2015
|
|
6HX9
 
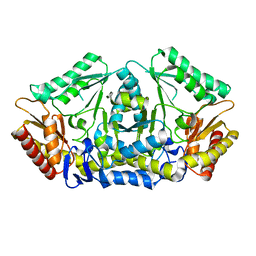 | | Putrescine transaminase from Pseudomonas putida | | 分子名称: | Aspartate aminotransferase family protein | | 著者 | Gahloth, D. | | 登録日 | 2018-10-16 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Characterization of a Putrescine Transaminase FromPseudomonas putidaand its Application to the Synthesis of Benzylamine Derivatives.
Front Bioeng Biotechnol, 6, 2018
|
|
3B43
 
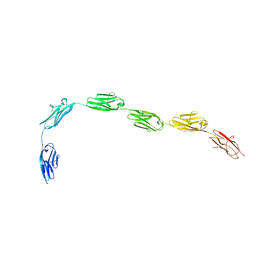 | | I-band fragment I65-I70 from titin | | 分子名称: | Titin | | 著者 | von Castelmur, E, Marino, M, Labeit, D, Labeit, S, Mayans, O. | | 登録日 | 2007-10-23 | | 公開日 | 2008-01-22 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | A regular pattern of Ig super-motifs defines segmental flexibility as the elastic mechanism of the titin chain
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4ZOT
 
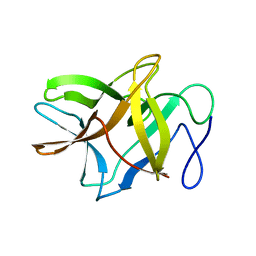 | | Crystal structure of BbKI, a disulfide-free plasma kallikrein inhibitor at 1.4 A resolution | | 分子名称: | Kunitz-type serine protease inhibitor BbKI | | 著者 | Shabalin, I.G, Zhou, D, Wlodawer, A, Oliva, M.L.V. | | 登録日 | 2015-05-06 | | 公開日 | 2015-05-20 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure of BbKI, a disulfide-free plasma kallikrein inhibitor.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
6H9C
 
 | | Cryo-EM structure of archaeal extremophilic internal membrane-containing Haloarcula californiae icosahedral virus 1 (HCIV-1) at 3.74 Angstroms resolution. | | 分子名称: | (Half) GPS-II protein located underneath the two-tower capsomer sitting ON the icosahedral 2-fold axis., GPS-II protein located underneath the two-tower capsomer NOT sitting on the icosahedral 2-fold axis., GPS-III molecule located underneath the capsomer close to the icosahedral three-fold axis., ... | | 著者 | Abrescia, N.G, Santos-Perez, I, Charro, D. | | 登録日 | 2018-08-03 | | 公開日 | 2019-03-27 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.74 Å) | | 主引用文献 | Structural basis for assembly of vertical single beta-barrel viruses.
Nat Commun, 10, 2019
|
|
8DF1
 
 | | Chi3l1 bound by antibody C59 | | 分子名称: | C59 Fab Heavy chain, C59 Fab Light chain, Chitinase-3-like protein 1, ... | | 著者 | Wrapp, D, McLellan, J.S. | | 登録日 | 2022-06-21 | | 公開日 | 2023-03-01 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | A novel humanized Chi3l1 blocking antibody attenuates acetaminophen-induced liver injury in mice.
Antib Ther, 6, 2023
|
|
8DEJ
 
 | | D. vulgaris type I-C Cascade bound to dsDNA target | | 分子名称: | CRISPR-associated protein, CT1133 family, TM1801 family, ... | | 著者 | O'Brien, R.E, Bravo, J.P.K, Ramos, D, Hibshman, G.N, Wright, J.T, Taylor, D.W. | | 登録日 | 2022-06-20 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.86 Å) | | 主引用文献 | Structural snapshots of R-loop formation by a type I-C CRISPR Cascade.
Mol.Cell, 83, 2023
|
|
6HL4
 
 | | wild-type NuoEF from Aquifex aeolicus - reduced form | | 分子名称: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | 著者 | Gerhardt, S, Friedrich, T, Einsle, O, Gnandt, E, Schulte, M, Fiegen, D. | | 登録日 | 2018-09-10 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
6HLI
 
 | | wild-type NuoEF from Aquifex aeolicus - reduced form bound to NAD+ | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | 著者 | Gerhardt, S, Friedrich, T, Einsle, O, Gnandt, E, Schulte, M, Fiegen, D. | | 登録日 | 2018-09-11 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
6HMC
 
 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 gene product) IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR THN27 | | 分子名称: | 1,2-ETHANEDIOL, 5-propan-2-yl-4-prop-2-enoxy-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, Casein kinase II subunit alpha' | | 著者 | Niefind, K, Lindenblatt, D, Dimper, V, Jose, J, Le Borgne, M. | | 登録日 | 2018-09-12 | | 公開日 | 2019-03-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.03 Å) | | 主引用文献 | Diacritic Binding of an Indenoindole Inhibitor by CK2 alpha Paralogs Explored by a Reliable Path to Atomic Resolution CK2 alpha ' Structures.
Acs Omega, 4, 2019
|
|
3K8E
 
 | | Crystal structure of E. coli lipopolysaccharide specific CMP-KDO synthetase | | 分子名称: | 3-deoxy-manno-octulosonate cytidylyltransferase | | 著者 | Heyes, D.J, Levy, C.W, Lafite, P, Scrutton, N.S, Leys, D. | | 登録日 | 2009-10-14 | | 公開日 | 2009-11-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Structure-based mechanism of CMP-2-keto-3-deoxymanno-octulonic acid synthetase: convergent evolution of a sugar-activating enzyme with DNA/RNA polymerases
J.Biol.Chem., 284, 2009
|
|
6J2J
 
 | | Crystal structure of bat (Pteropus Alecto) MHC class I Ptal-N*01:01 in complex with MERS-CoV-derived peptide MERS-CoV-S3 | | 分子名称: | Beta-2-microglobulin, MERS-CoV-S3, Ptal-N*01:01 | | 著者 | Lu, D, Liu, K.F, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, G.F, Liu, W.J. | | 登録日 | 2019-01-01 | | 公開日 | 2019-09-18 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
6B9W
 
 | |
6BAM
 
 | |
6BMB
 
 | | Crystal structure of Arabidopsis Dehydroquinate dehydratase-shikimate dehydrogenase (T381G mutant) in complex with tartrate and shikimate | | 分子名称: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic, ... | | 著者 | Christendat, D, Peek, J. | | 登録日 | 2017-11-14 | | 公開日 | 2017-12-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.077 Å) | | 主引用文献 | Structural and biochemical approaches uncover multiple evolutionary trajectories of plant quinate dehydrogenases.
Plant J., 2018
|
|
6BO7
 
 | | Crystal structure of Plasmodium vivax hypoxanthine guanine phosphoribosyltransferase in complex with [3R,4R]-4-guanin-9-yl-3-((S)-2-hydroxy-2-phosphonoethyl)oxy-1-N-(phosphonopropionyl)pyrrolidine | | 分子名称: | Hypoxanthine phosphoribosyltransferase, MAGNESIUM ION, [3-[(3~{R},4~{R})-3-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-4-[(2~{S})-2-oxidanyl-2-phosphono-ethoxy]pyrrolidin-1-y l]-3-oxidanylidene-propyl]phosphonic acid | | 著者 | Guddat, L.W, Keough, D.T, Rejman, D. | | 登録日 | 2017-11-18 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.856 Å) | | 主引用文献 | Design of Plasmodium vivax Hypoxanthine-Guanine Phosphoribosyltransferase Inhibitors as Potential Antimalarial Therapeutics.
ACS Chem. Biol., 13, 2018
|
|
5JM4
 
 | | Crystal structure of 14-3-3zeta in complex with a cyclic peptide involving an adamantyl and a dicarboxy side chain | | 分子名称: | 14-3-3 protein zeta/delta, BENZOIC ACID, GLN-GLY-MKD-ANG-ASP-MKD-LEU-ASP-LEU-ALA-CLU | | 著者 | Bier, D, Krueger, D, Glas, A, Wallraven, K, Ottmann, C, Hennig, S, Grossmann, T. | | 登録日 | 2016-04-28 | | 公開日 | 2017-05-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Structure-Based Design of Non-natural Macrocyclic Peptides That Inhibit Protein-Protein Interactions.
J. Med. Chem., 60, 2017
|
|
6ZS5
 
 | | 3.5 A cryo-EM structure of human uromodulin filament core | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Stanisich, J.J, Zyla, D, Afanasyev, P, Xu, J, Pilhofer, M, Boeringer, D, Glockshuber, R. | | 登録日 | 2020-07-15 | | 公開日 | 2020-09-02 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | The cryo-EM structure of the human uromodulin filament core reveals a unique assembly mechanism.
Elife, 9, 2020
|
|
6I6Z
 
 | |
8CYK
 
 | | Crystal structure of hallucinated protein HALC1_878 | | 分子名称: | HALC1_878 | | 著者 | Ragotte, R.J, Bera, A.K, Milles, L.F, Wicky, B.I.M, Baker, D. | | 登録日 | 2022-05-23 | | 公開日 | 2022-09-28 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Robust deep learning-based protein sequence design using ProteinMPNN.
Science, 378, 2022
|
|
7C04
 
 | | Crystal structure of human Trap1 with DN203492 | | 分子名称: | 4-chloranyl-1-[[2-methoxy-4-(trifluoromethyl)phenyl]methyl]pyrazolo[3,4-d]pyrimidin-6-amine, Heat shock protein 75 kDa, mitochondrial | | 著者 | Kim, D, Kim, D, Kim, S.Y, Lee, J.H, Kang, B.H, Kang, S, Lee, C. | | 登録日 | 2020-04-30 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Development of pyrazolo[3,4-d]pyrimidine-6-amine-based TRAP1 inhibitors that demonstrate in vivo anticancer activity in mouse xenograft models.
Bioorg.Chem., 101, 2020
|
|
7C05
 
 | | Crystal structure of human Trap1 with DN203495 | | 分子名称: | 1-[(4-bromanyl-2-fluoranyl-phenyl)methyl]-4-chloranyl-pyrazolo[3,4-d]pyrimidin-6-amine, Heat shock protein 75 kDa, mitochondrial | | 著者 | Kim, D, Kim, D, Kim, S.Y, Lee, J.H, Kang, B.H, Kang, S, Lee, C. | | 登録日 | 2020-04-30 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Development of pyrazolo[3,4-d]pyrimidine-6-amine-based TRAP1 inhibitors that demonstrate in vivo anticancer activity in mouse xenograft models.
Bioorg.Chem., 101, 2020
|
|
7TLY
 
 | |
1NX3
 
 | | Calpain Domain VI in Complex with the Inhibitor PD150606 | | 分子名称: | 3-(4-IODO-PHENYL)-2-MERCAPTO-PROPIONIC ACID, CALCIUM ION, Calcium-dependent protease, ... | | 著者 | Todd, B, Moore, D, Deivanayagam, C.C.S, Lin, G.-D, Chattopadhyay, D, Maki, M, Wang, K.K.W, Narayana, S.V.L. | | 登録日 | 2003-02-07 | | 公開日 | 2003-08-19 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | A structural model for the inhibition of calpain by calpastatin: crystal structures of the native domain VI of calpain and its complexes with calpastatin peptide and a small molecule inhibitor.
J.Mol.Biol., 328, 2003
|
|
6BW3
 
 | | Crystal structure of RBBP4 in complex with PRDM3 N-terminal peptide | | 分子名称: | Histone-binding protein RBBP4, MDS1 and EVI1 complex locus protein MDS1, UNKNOWN ATOM OR ION | | 著者 | Ivanochko, D, Halabelian, L, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | 登録日 | 2017-12-14 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Direct interaction between the PRDM3 and PRDM16 tumor suppressors and the NuRD chromatin remodeling complex.
Nucleic Acids Res., 47, 2019
|
|
