6DSI
 
 | |
6DQH
 
 | | Cronobacter sakazakii (Enterobacter sakazakii) Metallo-beta-lactamse HARLDQ motif | | Descriptor: | Beta-lactamase, PHOSPHATE ION, ZINC ION | | Authors: | Monteiro Pedroso, M, Waite, D, Natasa, M, McGeary, R, Guddat, L, Hugenholtz, P, Schenk, G. | | Deposit date: | 2018-06-11 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.104 Å) | | Cite: | Broad spectrum antibiotic-degrading metallo-beta-lactamases are phylogenetically diverse
Protein Cell, 2020
|
|
6U3Y
 
 | | Structure-based discovery of a novel small-molecule inhibitor of methicillin-resistant S. aureus | | Descriptor: | ACETATE ION, Gamma-hemolysin subunit A, Panton-Valentine Leucocidin F, ... | | Authors: | Liu, J, Kozhaya, L, Torres, V.J, Unutmaz, D, Lu, M. | | Deposit date: | 2019-08-22 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure-based discovery of a small-molecule inhibitor of methicillin-resistantStaphylococcus aureusvirulence.
J.Biol.Chem., 295, 2020
|
|
6U07
 
 | | Computational Stabilization of T Cell Receptor Constant Domains | | Descriptor: | MAGNESIUM ION, Stabilized T cell receptor constant domain (Calpha), Stabilized T cell receptor constant domain (Cbeta) | | Authors: | Froning, K, Maguire, J, Sereno, A, Huang, F, Chang, S, Weichert, K, Frommelt, A.J, Dong, J, Wu, X, Austin, H, Conner, E.M, Fitchett, J.R, Heng, A.R, Balasubramaniam, D, Hilgers, M.T, Kuhlman, B, Demarest, S.J. | | Deposit date: | 2019-08-13 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Computational stabilization of T cell receptors allows pairing with antibodies to form bispecifics.
Nat Commun, 11, 2020
|
|
6U1Y
 
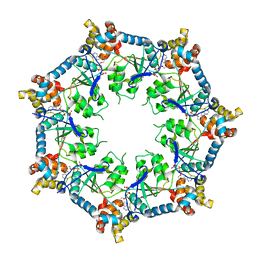 | | bcs1 AAA domain | | Descriptor: | MAGNESIUM ION, Mitochondrial chaperone BCS1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Tang, W.K, Xia, D. | | Deposit date: | 2019-08-17 | | Release date: | 2020-02-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structures of AAA protein translocase Bcs1 suggest translocation mechanism of a folded protein.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6U49
 
 | | Structure-based discovery of a novel small-molecule inhibitor of methicillin-resistant S. aureus | | Descriptor: | Alpha-hemolysin, SULFATE ION, fos-choline-14 | | Authors: | Liu, J, Kozhaya, L, Torres, V.J, Unutmaz, D, Lu, M. | | Deposit date: | 2019-08-23 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-based discovery of a small-molecule inhibitor of methicillin-resistantStaphylococcus aureusvirulence.
J.Biol.Chem., 295, 2020
|
|
6D9H
 
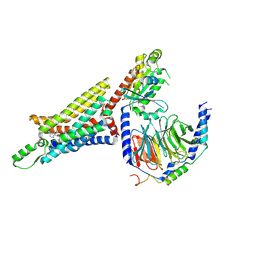 | | Cryo-EM structure of the human adenosine A1 receptor-Gi2-protein complex bound to its endogenous agonist | | Descriptor: | ADENOSINE, Chimera protein of Muscarinic acetylcholine receptor M4 and Adenosine receptor A1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Draper-Joyce, C.J, Khoshouei, M, Thal, D.M, Liang, Y.-L, Nguyen, A.T.N, Furness, S.G.B, Venugopal, H, Baltos, J, Plitzko, J.M, Danev, R, Baumeister, W, May, L.T, Wootten, D, Sexton, P, Glukhova, A, Christopoulos, A. | | Deposit date: | 2018-04-29 | | Release date: | 2018-06-20 | | Last modified: | 2018-07-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the adenosine-bound human adenosine A1receptor-Gicomplex.
Nature, 558, 2018
|
|
6U5J
 
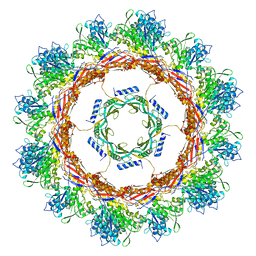 | | CryoEM Structure of Pyocin R2 - postcontracted - collar | | Descriptor: | Collar PA0615, Sheath PA0622 | | Authors: | Ge, P, Avaylon, J, Scholl, D, Shneider, M.M, Browning, C, Buth, S.A, Plattner, M, Ding, K, Leiman, P.G, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2019-08-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Action of a minimal contractile bactericidal nanomachine.
Nature, 580, 2020
|
|
6DDW
 
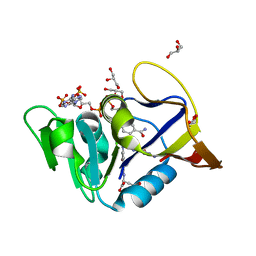 | | Mycobacterium tuberculosis Dihydrofolate Reductase complexed with beta-NADPH and N-(4-{[(2-amino-4-oxo-3,4-dihydropteridin-6-yl)methyl]amino}-2-hydroxybenzene-1-carbonyl)-L-glutamic acid | | Descriptor: | Dihydrofolate reductase, GLYCEROL, N-(4-{[(2-amino-4-oxo-3,4-dihydropteridin-6-yl)methyl]amino}-2-hydroxybenzene-1-carbonyl)-L-glutamic acid, ... | | Authors: | Hajian, B, Scocchera, E, Wright, D. | | Deposit date: | 2018-05-10 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Drugging the Folate Pathway in Mycobacterium tuberculosis: The Role of Multi-targeting Agents.
Cell Chem Biol, 26, 2019
|
|
6UCN
 
 | | Multi-conformer model of Ketosteroid Isomerase from Pseudomonas Putida (pKSI) bound to Equilenin at 250 K | | Descriptor: | CHLORIDE ION, EQUILENIN, MAGNESIUM ION, ... | | Authors: | Yabukarski, F, Herschlag, D, Biel, J.T, Fraser, J.S. | | Deposit date: | 2019-09-16 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Assessment of enzyme active site positioning and tests of catalytic mechanisms through X-ray-derived conformational ensembles.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6DG5
 
 | | Structure of a de novo designed Interleukin-2/Interleukin-15 mimetic complex with IL-2Rb and IL-2Rg | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit gamma, ... | | Authors: | Jude, K.M, Silva, D.-A, Yu, S, Baker, D, Garcia, K.C. | | Deposit date: | 2018-05-16 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.516 Å) | | Cite: | De novo design of potent and selective mimics of IL-2 and IL-15.
Nature, 565, 2019
|
|
6UF7
 
 | | S2 symmetric peptide design number 5, Uncle Fester | | Descriptor: | S2-5, Uncle Fester | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-23 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UF8
 
 | | S2 symmetric peptide design number 6, London Bridge | | Descriptor: | S2-6, London Bridge | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-23 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UFU
 
 | | C2 symmetric peptide design number 1, Zappy, crystal form 1 | | Descriptor: | C2-1, Zappy, crystal form 1 | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-25 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UG3
 
 | | C3 symmetric peptide design number 1, Sporty, crystal form 1 | | Descriptor: | C3-1, Sporty, crystal form 1, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-25 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UGC
 
 | | C3 symmetric peptide design number 3 | | Descriptor: | C3-3 cyclic peptide design, CADMIUM ION, SODIUM ION | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UDR
 
 | | S2 symmetric peptide design number 3 crystal form 1, Lurch | | Descriptor: | S2-3, Lurch crystal form 1 | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-23 | | Last modified: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6DLD
 
 | | Crystal structure of IgLON5/NEGR1 heterodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IgLON family member 5, Neuronal growth regulator 1, ... | | Authors: | Ranaivoson, F.M, Turk, L.S, Ozkan, E, Montelione, G.T, Comoletti, D. | | Deposit date: | 2018-06-01 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of a heterodimer of neuronal cell surface proteins
Structure, 2019
|
|
6UKS
 
 | | ATPgammaS bound mBcs1 | | Descriptor: | MAGNESIUM ION, Mitochondrial chaperone BCS1, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Tang, W.K, Borgnia, M.J, Hsu, A.L, Xia, D. | | Deposit date: | 2019-10-05 | | Release date: | 2020-02-05 | | Last modified: | 2020-02-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of AAA protein translocase Bcs1 suggest translocation mechanism of a folded protein.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6DJH
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ515 | | Descriptor: | 1,2-ETHANEDIOL, 8-bromo-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-05-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.918 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
6DQ2
 
 | | Cronobacter sakazakii (Enterobacter sakazakii) Metallo-beta-lactamse HARLDQ motif mutant S60 | | Descriptor: | Beta-lactamase, PHOSPHATE ION | | Authors: | Monteiro Pedroso, M, Waite, D, Natasa, M, McGeary, R, Guddat, L, Hugenholtz, P, Schenk, G. | | Deposit date: | 2018-06-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.158 Å) | | Cite: | Broad spectrum antibiotic-degrading metallo-beta-lactamases are phylogenetically diverse
Protein Cell, 2020
|
|
6U8D
 
 | | Crystal structure of hepatitis C virus IRES junction IIIabc in complex with Fab HCV2 | | Descriptor: | Heavy chain of Fab HCV2, JIIIabc RNA (68-MER), Light chain of Fab HCV2 | | Authors: | Koirala, D, Lewicka, A, Koldobskaya, Y, Huang, H, Piccirilli, J.A. | | Deposit date: | 2019-09-04 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.807 Å) | | Cite: | Synthetic Antibody Binding to a Preorganized RNA Domain of Hepatitis C Virus Internal Ribosome Entry Site Inhibits Translation.
Acs Chem.Biol., 15, 2020
|
|
6DRX
 
 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5HT2B receptor, BRIL chimera, ... | | Authors: | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DS0
 
 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | Descriptor: | (1S)-1-[(2-chloro-3,4-dimethoxyphenyl)methyl]-6-methyl-2,3,4,9-tetrahydro-1H-beta-carboline, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5HT2B receptor, ... | | Authors: | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.188 Å) | | Cite: | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DSL
 
 | | Consensus engineered intein (Cat) with atypical split site | | Descriptor: | Consensus engineered intein CatC, Consensus engineered intein CatN | | Authors: | Sekar, G, Stevens, A.J, Muir, T.W, Cowburn, D. | | Deposit date: | 2018-06-14 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | An Atypical Mechanism of Split Intein Molecular Recognition and Folding.
J. Am. Chem. Soc., 140, 2018
|
|
