4PK7
 
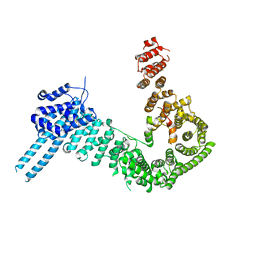 | | crystal structure of human Stromal Antigen 2 (SA2) in complex with Sister Chromatid Cohesion protein 1 (Scc1) with bound MES, native proteins | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cohesin subunit SA-2, Double-strand-break repair protein rad21 homolog | | Authors: | Hara, K, Chen, Z, Tomchick, D.R, Yu, H. | | Deposit date: | 2014-05-13 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of cohesin subcomplex pinpoints direct shugoshin-Wapl antagonism in centromeric cohesion.
Nat.Struct.Mol.Biol., 21, 2014
|
|
5O51
 
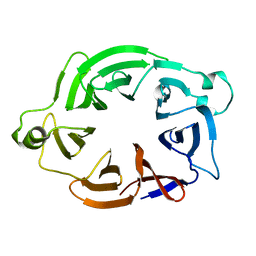 | | AfRom2 CNH domain | | Descriptor: | Rho guanyl nucleotide exchange factor (Rom2), putative | | Authors: | Wei, W, van Aalten, D. | | Deposit date: | 2017-05-31 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Rom2 CNH domain from Aspergillus fumigatus is an atypical seven-bladed WD-40 protein
To Be Published
|
|
1Z3H
 
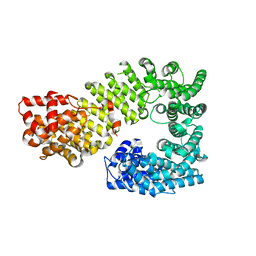 | | The exportin Cse1 in its cargo-free, cytoplasmic state | | Descriptor: | Importin alpha re-exporter, MAGNESIUM ION | | Authors: | Cook, A, Fernandez, E, Lindner, D, Ebert, J, Schlenstedt, G, Conti, E. | | Deposit date: | 2005-03-12 | | Release date: | 2005-05-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of the nuclear export receptor cse1 in its cytosolic state reveals a closed conformation incompatible with cargo binding
Mol.Cell, 18, 2005
|
|
8F0E
 
 | |
4YPT
 
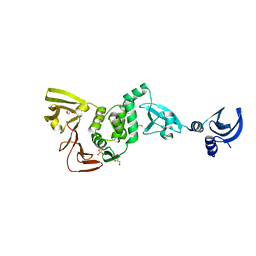 | | X-ray structural of three tandemly linked domains of nsp3 from murine hepatitis virus at 2.60 Angstroms resolution | | Descriptor: | GLYCEROL, Replicase polyprotein 1ab, ZINC ION | | Authors: | Chen, Y, Savinov, S.N, Mielech, A.M, Cao, T, Baker, S.C, Mesecar, A.D. | | Deposit date: | 2015-03-13 | | Release date: | 2015-08-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6009 Å) | | Cite: | X-ray Structural and Functional Studies of the Three Tandemly Linked Domains of Non-structural Protein 3 (nsp3) from Murine Hepatitis Virus Reveal Conserved Functions.
J.Biol.Chem., 290, 2015
|
|
2XKV
 
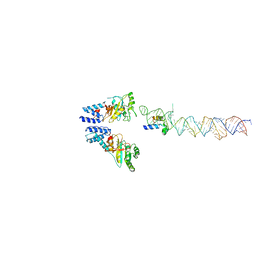 | | Atomic Model of the SRP-FtsY Early Conformation | | Descriptor: | 4.5S RNA, CELL DIVISION PROTEIN FTSY, SIGNAL RECOGNITION PARTICLE PROTEIN | | Authors: | Estrozi, L.F, Boehringer, D, Shan, S.-o, Ban, N, Schaffitzel, C. | | Deposit date: | 2010-07-13 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Cryo-Em Structure of the E. Coli Translating Ribosome in Complex with Srp and its Receptor.
Nat.Struct.Mol.Biol., 18, 2011
|
|
8C4I
 
 | |
3ZXX
 
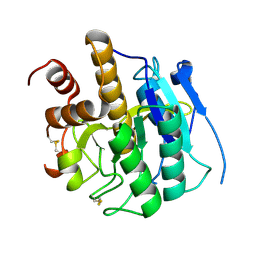 | | Structure of self-cleaved protease domain of PatA | | Descriptor: | SUBTILISIN-LIKE PROTEIN | | Authors: | Koehnke, J, Zollman, D, Vendome, J, Raab, A, Houssen, W.E, Smith, M.C, Jaspars, M, Naismith, J.H. | | Deposit date: | 2011-08-16 | | Release date: | 2012-08-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Discovery of New Cyanobactins from Cyanothece Pcc 7425 Defines a New Signature for Processing of Patellamides.
Chembiochem, 13, 2012
|
|
6IDY
 
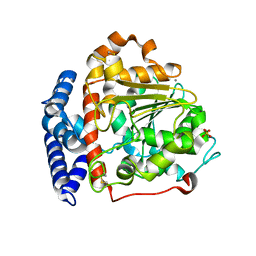 | |
5JU7
 
 | | DNA BINDING DOMAIN OF E.COLI CADC | | Descriptor: | Transcriptional activator CadC, ZINC ION | | Authors: | Janowski, R, Schlundt, A, Sattler, M, Niessing, D. | | Deposit date: | 2016-05-10 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-function analysis of the DNA-binding domain of a transmembrane transcriptional activator.
Sci Rep, 7, 2017
|
|
2FGG
 
 | | Crystal Structure of Rv2632c | | Descriptor: | Hypothetical protein Rv2632c/MT2708 | | Authors: | Yu, M, Bursey, E.H, Radhakannan, T, Segelke, B.W, Lekin, T, Toppani, D, Kim, C.Y, Kaviratne, T, Woodruff, T, Terwilliger, T.C, Hung, L.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-12-21 | | Release date: | 2006-02-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Rv2632c
To be Published
|
|
9AXE
 
 | |
6IJH
 
 | | Crystal structure of PDE10 in complex with inhibitor AF-399/14387019 | | Descriptor: | 2-[2-(4-phenyl-5-sulfanylidene-4,5-dihydro-1H-1,2,4-triazol-3-yl)ethyl]-1H-benzo[de]isoquinoline-1,3(2H)-dione, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huang, Y.Y, Yu, Y.F, Zhang, C, Wu, D, Wu, Y, Luo, H.B. | | Deposit date: | 2018-10-10 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Validation of Phosphodiesterase-10 as a Novel Target for Pulmonary Arterial Hypertension via Highly Selective and Subnanomolar Inhibitors.
J. Med. Chem., 62, 2019
|
|
8OPQ
 
 | | Structure of Human Solute Carrier 26 family member A6 (SLC26A6) anion transporter in an inward-facing state | | Descriptor: | CHLORIDE ION, Solute carrier family 26 member 6 | | Authors: | Tippett, D.N, Breen, C, Butler, S.J, Sawicka, M, Dutzler, R. | | Deposit date: | 2023-04-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural and functional properties of the transporter SLC26A6 reveal mechanism of coupled anion exchange.
Elife, 12, 2023
|
|
4X5U
 
 | | X-ray crystal structure of CagL at pH 4.2 | | Descriptor: | Cag pathogenicity island protein (Cag18) | | Authors: | Sundberg, E.J, Bonsor, D.A, Diederichs, K. | | Deposit date: | 2014-12-05 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Integrin Engagement by the Helical RGD Motif of the Helicobacter pylori CagL Protein Is Regulated by pH-induced Displacement of a Neighboring Helix.
J.Biol.Chem., 290, 2015
|
|
1KUL
 
 | | GLUCOAMYLASE, GRANULAR STARCH-BINDING DOMAIN, NMR, 5 STRUCTURES | | Descriptor: | GLUCOAMYLASE | | Authors: | Sorimachi, K, Jacks, A.J, Le Gal-Coeffet, M.-F, Williamson, G, Archer, D.B, Williamson, M.P. | | Deposit date: | 1996-01-12 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the granular starch binding domain of glucoamylase from Aspergillus niger by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 259, 1996
|
|
6W7P
 
 | |
1KUN
 
 | | SOLUTION STRUCTURE OF THE HUMAN ALPHA3-CHAIN TYPE VI COLLAGEN C-TERMINAL KUNITZ DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | ALPHA3-CHAIN TYPE VI COLLAGEN | | Authors: | Sorensen, M.D, Bjorn, S, Norris, K, Olsen, O, Petersen, L, James, T.L, Led, J.J. | | Deposit date: | 1997-03-04 | | Release date: | 1997-11-12 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the human alpha3-chain type VI collagen C-terminal Kunitz domain,.
Biochemistry, 36, 1997
|
|
4WF8
 
 | | Crystal structure of NS3/4A protease in complex with Asunaprevir | | Descriptor: | CHLORIDE ION, N-(tert-butoxycarbonyl)-3-methyl-L-valyl-(4R)-4-[(7-chloro-4-methoxyisoquinolin-1-yl)oxy]-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-L-prolinamide, NS3 protein, ... | | Authors: | Schiffer, C.A, Soumana, D.I, Ali, A. | | Deposit date: | 2014-09-13 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Analysis of Asunaprevir Resistance in HCV NS3/4A Protease.
Acs Chem.Biol., 9, 2014
|
|
1KUM
 
 | | GLUCOAMYLASE, GRANULAR STARCH-BINDING DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GLUCOAMYLASE | | Authors: | Sorimachi, K, Jacks, A.J, Le Gal-Coeffet, M.-F, Williamson, G, Archer, D.B, Williamson, M.P. | | Deposit date: | 1996-01-12 | | Release date: | 1996-07-11 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the granular starch binding domain of glucoamylase from Aspergillus niger by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 259, 1996
|
|
7XX1
 
 | | Crystal structure of SARS-CoV-2 N-NTD | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nucleoprotein, ZINC ION | | Authors: | Luan, X.D, Li, X.M, Li, Y.F. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antiviral drug design based on structural insights into the N-terminal domain and C-terminal domain of the SARS-CoV-2 nucleocapsid protein.
Sci Bull (Beijing), 67, 2022
|
|
2WV9
 
 | | Crystal Structure of the NS3 protease-helicase from Murray Valley encephalitis virus | | Descriptor: | FLAVIVIRIN PROTEASE NS2B REGULATORY SUBUNIT, FLAVIVIRIN PROTEASE NS3 CATALYTIC SUBUNIT | | Authors: | Assenberg, R, Mastrangelo, E, Walter, T.S, Verma, A, Milani, M, Owens, R.J, Stuart, D.I, Grimes, J.M, Mancini, E.J. | | Deposit date: | 2009-10-15 | | Release date: | 2009-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of a Novel Conformational State of the Flavivirus Ns3 Protein: Implications for Polyprotein Processing and Viral Replication.
J.Virol., 83, 2009
|
|
6W25
 
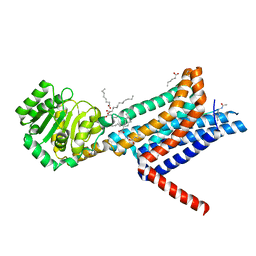 | | Crystal structure of the Melanocortin-4 Receptor (MC4R) in complex with SHU9119 | | Descriptor: | CALCIUM ION, Melanocortin receptor 4,GlgA glycogen synthase,Melanocortin receptor 4, OLEIC ACID, ... | | Authors: | Yu, J, Gimenez, L.E, Hernandez, C.C, Wu, Y, Wein, A.H, Han, G.W, McClary, K, Mittal, S.R, Burdsall, K, Stauch, B, Wu, L, Stevens, S.N, Peisley, A, Williams, S.Y, Chen, V, Millhauser, G.L, Zhao, S, Cone, R.D, Stevens, R.C. | | Deposit date: | 2020-03-04 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Determination of the melanocortin-4 receptor structure identifies Ca2+as a cofactor for ligand binding.
Science, 368, 2020
|
|
4WIL
 
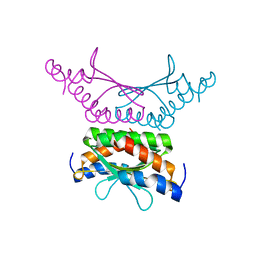 | | Crystal structure of DCoH2 S51T | | Descriptor: | Pterin-4-alpha-carbinolamine dehydratase 2 | | Authors: | Wang, D, Rose, R.B. | | Deposit date: | 2014-09-26 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Interactions with the Bifunctional Interface of the Transcriptional Coactivator DCoH1 Are Kinetically Regulated.
J.Biol.Chem., 290, 2015
|
|
4WIS
 
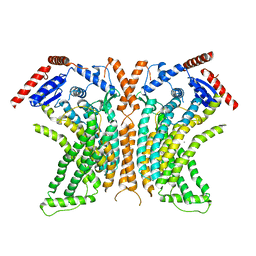 | | Crystal structure of the lipid scramblase nhTMEM16 in crystal form 1 | | Descriptor: | CALCIUM ION, lipid scramblase | | Authors: | Dutzler, R, Brunner, J.D, Lim, N.K, Schenck, S. | | Deposit date: | 2014-09-26 | | Release date: | 2014-11-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | X-ray structure of a calcium-activated TMEM16 lipid scramblase.
Nature, 516, 2014
|
|
