6BE7
 
 | | Solution structure of de novo macrocycle Design8.1 | | Descriptor: | DDPT(DPR)(DAR)Q(DGN) | | Authors: | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | Deposit date: | 2017-10-24 | | Release date: | 2018-01-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
7B2N
 
 | | Crystal structure of Chlamydomonas reinhardtii chloroplastic Fructose bisphosphate aldolase | | Descriptor: | CHLORIDE ION, Fructose-bisphosphate aldolase 1, chloroplastic, ... | | Authors: | Le Moigne, T, Lemaire, S.D, Henri, J. | | Deposit date: | 2020-11-27 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structure of chloroplast fructose-1,6-bisphosphate aldolase from the green alga Chlamydomonas reinhardtii.
J.Struct.Biol., 214, 2022
|
|
6BEN
 
 | | Solution structure of de novo macrocycle design8.2 | | Descriptor: | (DAR)Q(DPR)(DGN)R(DGL)PQ | | Authors: | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, B. | | Deposit date: | 2017-10-25 | | Release date: | 2017-12-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
7QCF
 
 | |
5EFJ
 
 | |
8CDM
 
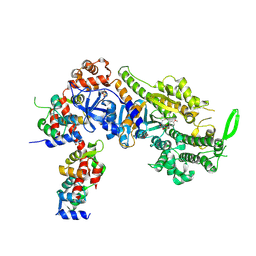 | | Plasmodium falciparum Myosin A full-length, post-rigor state complexed to the inhibitor KNX-002 | | Descriptor: | 1,2-ETHANEDIOL, 1-(4-methoxyphenyl)-~{N}-[(3-thiophen-2-yl-1~{H}-pyrazol-4-yl)methyl]cyclopropan-1-amine, Myosin A tail domain interacting protein, ... | | Authors: | Moussaoui, D, Robblee, J.P, Robert-Paganin, J, Auguin, D, Fisher, F, Fagnant, P.M, MacFarlane, J.E, Mueller-Dieckmann, C, Baum, J, Trybus, K.M, Houdusse, A. | | Deposit date: | 2023-01-31 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of small molecule inhibition of Plasmodium falciparum myosin A informs antimalarial drug design.
Nat Commun, 14, 2023
|
|
5U38
 
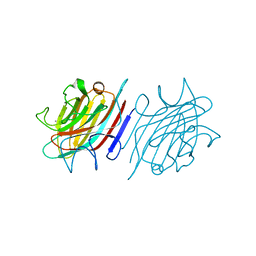 | | Crystal structure of native lectin from Platypodium elegans seeds (PELa) complexed with Man1-3Man-OMe. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Lectin, ... | | Authors: | Silva, I.B, Araripe, D.A, Neco, A.H.B, Pinto-Junior, V.R, Osterne, V.J.S, Santiago, M.Q, Silva-Filho, J.C, Leal, R.B, Rocha, C.R.C, Nascimento, K.S, Cavada, B.S. | | Deposit date: | 2016-12-01 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural studies and nociceptive activity of a native lectin from Platypodium elegans seeds (nPELa).
Int. J. Biol. Macromol., 107, 2018
|
|
7ZGU
 
 | | Human NLRP3-deltaPYD hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NACHT, LRR and PYD domains-containing protein 3 | | Authors: | Raisch, T, Machtens, D.A, Bresch, I.B, Eberhage, J, Prumbaum, D, Reubold, T.F, Raunser, S, Eschenburg, S. | | Deposit date: | 2022-04-04 | | Release date: | 2022-05-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the NEK7-independent NLRP3 inflammasome
To Be Published
|
|
8CGQ
 
 | |
8PS0
 
 | |
6F0V
 
 | | GLIC mutant E82Q | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Hu, H.D, Delarue, M. | | Deposit date: | 2017-11-20 | | Release date: | 2018-01-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Full mutational mapping of titratable residues helps to identify proton-sensors involved in the control of channel gating in the Gloeobacter violaceus pentameric ligand-gated ion channel.
PLoS Biol., 15, 2017
|
|
4YAC
 
 | | Crystal structure of LigO in complex with NADH from Sphingobium sp. strain SYK-6 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, C alpha-dehydrogenase | | Authors: | Pereira, J.H, McAndrew, R.P, Heins, R.A, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structural and Biochemical Characterization of the Early and Late Enzymes in the Lignin beta-Aryl Ether Cleavage Pathway from Sphingobium sp. SYK-6.
J.Biol.Chem., 291, 2016
|
|
6F11
 
 | | GLIC mutant D86A | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Hu, H.D, Delarue, M. | | Deposit date: | 2017-11-21 | | Release date: | 2018-01-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Full mutational mapping of titratable residues helps to identify proton-sensors involved in the control of channel gating in the Gloeobacter violaceus pentameric ligand-gated ion channel.
PLoS Biol., 15, 2017
|
|
8A09
 
 | | A hexameric barrel state of a de novo coiled-coil assembly: CC-Type2-(QgLaId)4 | | Descriptor: | A hexameric barrel state of a de novo coiled-coil assembly: CC-Type2-(QgLaId)4, GLYCEROL | | Authors: | Martin, F.J.O, Rhys, G.G, Dawson, W.M, Woolfson, D.N. | | Deposit date: | 2022-05-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Differential sensing with arrays of de novo designed peptide assemblies.
Nat Commun, 14, 2023
|
|
6VHU
 
 | | Klebsiella oxytoca NpsA N-terminal subdomain in space group P21 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Kreitler, D.F, Gulick, A.M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biosynthesis, Mechanism of Action, and Inhibition of the Enterotoxin Tilimycin Produced by the Opportunistic PathogenKlebsiella oxytoca.
Acs Infect Dis., 6, 2020
|
|
6SC5
 
 | | dAb3/HOIP-RBR-Ligand2 | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF31, SULFATE ION, ... | | Authors: | Tsai, Y.-C.I, Johansson, H, House, D, Rittinger, K. | | Deposit date: | 2019-07-23 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Single-Domain Antibodies as Crystallization Chaperones to Enable Structure-Based Inhibitor Development for RBR E3 Ubiquitin Ligases.
Cell Chem Biol, 27, 2020
|
|
6BAR
 
 | | Crystal structure of Thermus thermophilus Rod shape determining protein RodA (Q5SIX3_THET8) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Rod shape determining protein RodA | | Authors: | Sjodt, M, Brock, K, Dobihal, G, Rohs, P.D.A, Green, A.G, Hopf, T.A, Meeske, A.J, Marks, D.S, Bernhardt, T.G, Rudner, D.Z, Kruse, A.C. | | Deposit date: | 2017-10-15 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis.
Nature, 556, 2018
|
|
4YT0
 
 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with 2-methyl-N-[3-(1-methylethoxy)phenyl]benzamide. | | Descriptor: | 2-methyl-N-[3-(1-methylethoxy)phenyl]benzamide, Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-03-17 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.66 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
6PDN
 
 | | Human PIM1 bound to benzothiophene inhibitor 292 | | Descriptor: | 4-{5-[(3-aminopropyl)carbamoyl]thiophen-2-yl}-1-benzothiophene-2-carboxylic acid, GLYCEROL, Serine/threonine-protein kinase pim-1, ... | | Authors: | Godoi, P.H.C, Santiago, A.S, Fala, A.M, Ramos, P.Z, Sriranganadane, D, Mascarello, A, Segretti, N, Azevedo, H, Guimaraes, C.R.W, Arruda, P, Elkins, J.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-19 | | Release date: | 2019-07-24 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human PIM1
To Be Published
|
|
4YHS
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM BRADYRHIZOBIUM sp. BTAi1 (BBta_2440, TARGET EFI-511490) WITH BOUND BIS-TRIS | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Monosaccharide ABC transporter substrate-binding protein, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-02-27 | | Release date: | 2015-03-18 | | Last modified: | 2015-10-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM BRADYRHIZOBIUM sp. BTAi1 (BBta_2440, TARGET EFI-511490) WITH BOUND BIS-TRIS
To be published
|
|
4YI9
 
 | |
6PDO
 
 | | Human PIM1 bound to benzothiophene inhibitor 354 | | Descriptor: | 4-{5-[(3-aminopropyl)carbamoyl]thiophen-2-yl}-1-benzothiophene-2-carboxamide, Peptide, Serine/threonine-protein kinase pim-1 | | Authors: | Godoi, P.H.C, Santiago, A.S, Fala, A.M, Ramos, P.Z, Sriranganadane, D, Mascarello, A, Segretti, N, Azevedo, H, Guimaraes, C.R.W, Arruda, P, Elkins, J.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-19 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human PIM1
To Be Published
|
|
8CMM
 
 | | Re-pairing DNA - binding of a ruthenium phi complex to a double mismatch | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*AP*TP*AP*AP*TP*GP*CP*G)-3'), LITHIUM ION, POTASSIUM ION, ... | | Authors: | Prieto Otoya, T.D, Cardin, C.J, McQuaid, K.M. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Re-pairing DNA: binding of a ruthenium phi complex to a double mismatch.
Chem Sci, 15, 2024
|
|
7AMG
 
 | | IL-17A in complex with small molecule modulators | | Descriptor: | (3~{R})-4-[4-[[(2~{S})-2-[[2,2-bis(fluoranyl)-2-phenyl-ethanoyl]amino]-3-(2-chlorophenyl)propanoyl]amino]phenyl]-3-[[(2~{S})-3-methyl-2-[2-[2-[(2-methylpropan-2-yl)oxycarbonylamino]ethoxy]ethanoylamino]butanoyl]amino]butanoic acid, Interleukin-17A | | Authors: | Hakansson, M, Kimbung, R, Logan, D, Walse, U.B, de Groot, M.J, Andrews, M.D, Dack, K.N, Lambert, M. | | Deposit date: | 2020-10-08 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Discovery of an Oral, Rule of 5 Compliant, Interleukin 17A Protein-Protein Interaction Modulator for the Potential Treatment of Psoriasis and Other Inflammatory Diseases.
J.Med.Chem., 65, 2022
|
|
8PM9
 
 | | Crystal structure of human wild type transthyretin in complex with PITB (Pharmacokinetically Improved TTR Binder) | | Descriptor: | (3-fluoranyl-5-oxidanyl-phenyl)-(3-methoxy-5-nitro-4-oxidanyl-phenyl)methanone, Transthyretin | | Authors: | Varejao, N, Pinheiro, F, Pallares, I, Ventura, S, Reverter, D. | | Deposit date: | 2023-06-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | PITB: A high affinity transthyretin aggregation inhibitor with optimal pharmacokinetic properties.
Eur.J.Med.Chem., 261, 2023
|
|
