5KLS
 
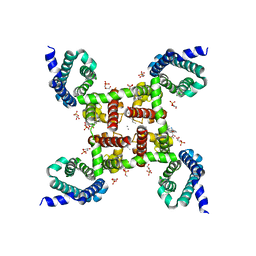 | | Structure of CavAb in complex with Br-dihydropyridine derivative UK-59811 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein, ... | | Authors: | Tang, L, Gamal EL-Din, T.M, Swanson, T.M, Pryde, D.C, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2016-06-25 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.299 Å) | | Cite: | Structural basis for inhibition of a voltage-gated Ca(2+) channel by Ca(2+) antagonist drugs.
Nature, 537, 2016
|
|
6PYE
 
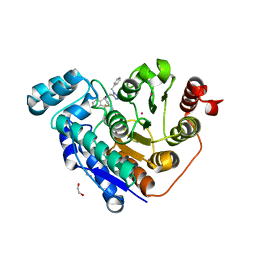 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with NR160 | | Descriptor: | 1,2-ETHANEDIOL, Hdac6 protein, N-[(1-benzyl-1H-tetrazol-5-yl)methyl]-N-{[4-(hydroxycarbamoyl)phenyl]methyl}-2-(trifluoromethyl)benzamide, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2019-07-29 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.480003 Å) | | Cite: | Multicomponent Synthesis, Binding Mode, and Structure-Activity Relationship of Selective Histone Deacetylase 6 (HDAC6) Inhibitors with Bifurcated Capping Groups.
J.Med.Chem., 63, 2020
|
|
4R6S
 
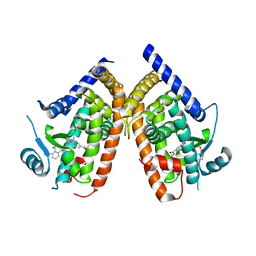 | | Crystal structure of PPARgammma in complex with SR1663 | | Descriptor: | 4'-[(2,3-dimethyl-5-{[(1R)-1-(4-nitrophenyl)ethyl]carbamoyl}-1H-indol-1-yl)methyl]biphenyl-2-carboxylic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Marciano, D.P, Griffin, P.R, Bruning, J.B. | | Deposit date: | 2014-08-26 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Pharmacological repression of PPAR gamma promotes osteogenesis.
Nat Commun, 6, 2015
|
|
2IE3
 
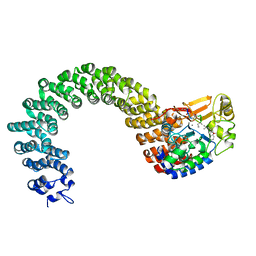 | | Structure of the Protein Phosphatase 2A Core Enzyme Bound to Tumor-inducing Toxins | | Descriptor: | MANGANESE (II) ION, Protein Phosphatase 2, regulatory subunit A (PR 65), ... | | Authors: | Xing, Y, Xu, Y, Chen, Y, Jeffrey, P.D, Chao, Y, Shi, Y. | | Deposit date: | 2006-09-17 | | Release date: | 2006-11-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Protein Phosphatase 2A Core Enzyme Bound to Tumor-Inducing Toxins
Cell(Cambridge,Mass.), 127, 2006
|
|
4RJK
 
 | | Acetolactate synthase from Bacillus subtilis bound to LThDP - crystal form II | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-CARBOXY-1-HYDROXYETHYL)-5-(2-{[HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, Acetolactate synthase, MAGNESIUM ION, ... | | Authors: | Sommer, B, von Moeller, H, Haack, M, Qoura, F, Langner, C, Bourenkov, G, Garbe, D, Brueck, T, Loll, B. | | Deposit date: | 2014-10-09 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Detailed Structure-Function Correlations of Bacillus subtilis Acetolactate Synthase.
Chembiochem, 16, 2015
|
|
8A4X
 
 | | Crystal structure of human Cathepsin L with covalently bound Calpain inhibitor III | | Descriptor: | (phenylmethyl) N-[(2S)-3-methyl-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-phenyl-propan-2-yl]amino]butan-2-yl]carbamate, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
1T0A
 
 | | Crystal Structure of 2C-Methyl-D-Erythritol-2,4-cyclodiphosphate Synthase from Shewanella Oneidensis | | Descriptor: | 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, COBALT (II) ION, FARNESYL DIPHOSPHATE, ... | | Authors: | Ni, S, Robinson, H, Marsing, G.C, Bussiere, D.E, Kennedy, M.A. | | Deposit date: | 2004-04-08 | | Release date: | 2004-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of 2C-methyl-D-erythritol-2,4-cyclodiphosphate synthase from Shewanella oneidensis at 1.6 A: identification of farnesyl pyrophosphate trapped in a hydrophobic cavity.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2ITM
 
 | | Crystal structure of the E. coli xylulose kinase complexed with xylulose | | Descriptor: | AMMONIUM ION, D-XYLULOSE, SULFATE ION, ... | | Authors: | di Luccio, E, Voegtli, J, Wilson, D.K. | | Deposit date: | 2006-10-19 | | Release date: | 2006-11-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and kinetic studies of induced fit in xylulose kinase from Escherichia coli.
J.Mol.Biol., 365, 2007
|
|
3ERT
 
 | | HUMAN ESTROGEN RECEPTOR ALPHA LIGAND-BINDING DOMAIN IN COMPLEX WITH 4-HYDROXYTAMOXIFEN | | Descriptor: | 4-HYDROXYTAMOXIFEN, PROTEIN (ESTROGEN RECEPTOR ALPHA) | | Authors: | Shiau, A.K, Barstad, D, Loria, P.M, Cheng, L, Kushner, P.J, Agard, D.A, Greene, G.L. | | Deposit date: | 1999-03-30 | | Release date: | 1999-04-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen.
Cell(Cambridge,Mass.), 95, 1998
|
|
6WCX
 
 | |
1TBH
 
 | | H141D mutant of rat liver arginase I | | Descriptor: | Arginase 1, MANGANESE (II) ION | | Authors: | Cama, E, Cox, J.D, Ash, D.E, Christianson, D.W. | | Deposit date: | 2004-05-20 | | Release date: | 2005-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Probing the role of the hyper-reactive histidine residue of arginase.
Arch.Biochem.Biophys., 444, 2005
|
|
4TRA
 
 | |
2IUT
 
 | | P. aeruginosa FtsK motor domain, dimeric | | Descriptor: | DNA TRANSLOCASE FTSK, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Massey, T.H, Mercogliano, C.P, Yates, J, Sherratt, D.J, Lowe, J. | | Deposit date: | 2006-06-07 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Double-Stranded DNA Translocation: Structure and Mechanism of Hexameric Ftsk
Mol.Cell, 23, 2006
|
|
7SUV
 
 | | APE1 exonuclease substrate complex with 8oxoG opposite A | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(8OG))-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Whitaker, A.W, Freudenthal, B.D. | | Deposit date: | 2021-11-18 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Processing oxidatively damaged bases at DNA strand breaks by APE1.
Nucleic Acids Res., 50, 2022
|
|
7SVB
 
 | | APE1 exonuclease substrate complex with 8oxoG opposite C | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(8OG))-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*CP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Whitaker, A.W, Freudenthal, B.D. | | Deposit date: | 2021-11-18 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Processing oxidatively damaged bases at DNA strand breaks by APE1.
Nucleic Acids Res., 50, 2022
|
|
1TKK
 
 | | The Structure of a Substrate-Liganded Complex of the L-Ala-D/L-Glu Epimerase from Bacillus subtilis | | Descriptor: | ALANINE, GLUTAMIC ACID, MAGNESIUM ION, ... | | Authors: | Klenchin, V.A, Schmidt, D.M, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-06-08 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolution of Enzymatic Activities in the Enolase Superfamily: Structure of a Substrate-Liganded Complex of the l-Ala-d/l-Glu Epimerase from Bacillus subtilis(,).
Biochemistry, 43, 2004
|
|
2IO3
 
 | | Crystal structure of human Senp2 in complex with RanGAP1-SUMO-2 | | Descriptor: | Ran GTPase-activating protein 1, Sentrin-specific protease 2, Small ubiquitin-related modifier 2 | | Authors: | Reverter, D, Lima, C.D. | | Deposit date: | 2006-10-09 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for SENP2 protease interactions with SUMO precursors and conjugated substrates.
Nat.Struct.Mol.Biol., 13, 2006
|
|
3F73
 
 | | Alignment of guide-target seed duplex within an argonaute silencing complex | | Descriptor: | ARGONAUTE, DNA (5'-D(P*DTP*DGP*DAP*DGP*DGP*DTP*DAP*DGP*DTP*DAP*DGP*DGP*DTP*DTP*DGP*DTP*DA*DTP*DAP*DGP*DT)-3'), MAGNESIUM ION, ... | | Authors: | Wang, Y, Li, H, Sheng, G, Juranek, S, Tuschl, T, Patel, D.J. | | Deposit date: | 2008-11-07 | | Release date: | 2008-12-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex.
Nature, 456, 2008
|
|
2VCE
 
 | | Characterization and engineering of the bifunctional N- and O- glucosyltransferase involved in xenobiotic metabolism in plants | | Descriptor: | 1,2-ETHANEDIOL, 2,4,5-trichlorophenol, HYDROQUINONE GLUCOSYLTRANSFERASE, ... | | Authors: | Brazier-Hicks, M, Offen, W.A, Gershater, M.C, Revett, T.J, Lim, E.K, Bowles, D.J, Davies, G.J, Edwards, R. | | Deposit date: | 2007-09-20 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization and Engineering of the Bifunctional N- and O-Glucosyltransferase Involved in Xenobiotic Metabolism in Plants.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
3WNE
 
 | |
7SLR
 
 | | HIV Reverse Transcriptase with compound Pyr01 | | Descriptor: | 5-(difluoromethyl)-3-({1-[(5-fluoro-2-oxo-1,2-dihydropyridin-3-yl)methyl]-6-oxo-4-(1,1,2,2-tetrafluoroethyl)-1,6-dihydropyrimidin-5-yl}oxy)-2-methylbenzonitrile, Reverse transcriptase/ribonuclease H | | Authors: | Klein, D.J, Zebisch, M, Gu, M. | | Deposit date: | 2021-10-24 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.179 Å) | | Cite: | Potent targeted activator of cell kill molecules eliminate cells expressing HIV-1.
Sci Transl Med, 15, 2023
|
|
1JPS
 
 | | Crystal structure of tissue factor in complex with humanized Fab D3h44 | | Descriptor: | immunoglobulin Fab D3H44, heavy chain, light chain, ... | | Authors: | Faelber, K, Kirchhofer, D, Presta, L, Kelley, R.F, Muller, Y.A. | | Deposit date: | 2001-08-03 | | Release date: | 2002-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The 1.85 A resolution crystal structures of tissue factor in complex with humanized Fab D3h44 and of free humanized Fab D3h44: revisiting the solvation of antigen combining sites.
J.Mol.Biol., 313, 2001
|
|
3EN9
 
 | | Structure of the Methanococcus jannaschii KAE1-BUD32 fusion protein | | Descriptor: | HEXATANTALUM DODECABROMIDE, MAGNESIUM ION, O-sialoglycoprotein endopeptidase/protein kinase | | Authors: | Neculai, D. | | Deposit date: | 2008-09-25 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine.
Mol.Cell, 32, 2008
|
|
5UMR
 
 | | Crystal structure of N-terminal domain of human FACT complex subunit SSRP1 | | Descriptor: | FACT complex subunit SSRP1 | | Authors: | Su, D, Hu, Q, Thompson, J.R, Heroux, A, Botuyan, M.V, Mer, G. | | Deposit date: | 2017-01-29 | | Release date: | 2018-01-31 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal structure of N-terminal domain of human FACT complex subunit SSRP1
To Be Published
|
|
6ISH
 
 | | Structure of 9N-I DNA polymerase incorporation with 3'-AL in the active site | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*CP*TP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*CP*CP*(DAL))-3'), ... | | Authors: | Linwu, S.W, Maestre-Reyna, M, Tsai, M.D, Tu, Y.H, Chang, W.H. | | Deposit date: | 2018-11-16 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Thermococcus sp. 9°N DNA polymerase exhibits 3'-esterase activity that can be harnessed for DNA sequencing.
Commun Biol, 2, 2019
|
|
