8DXI
 
 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment [1-(4-fluorophenyl)-5-methyl-1H-pyrazol-4-yl]methanol at multiple sites | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
8DXK
 
 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 4-bromopyrazole at multiple sites | | Descriptor: | 1,2-ETHANEDIOL, 4-bromo-1H-pyrazole, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
7NC7
 
 | | Crystal structure of fructose-bisphosphate aldolases FBAC from Bacillus methanolicus | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Fructose-bisphosphate aldolase | | Authors: | Einsle, O, Zhang, L, Guetle, D, Jacquot, J.P. | | Deposit date: | 2021-01-28 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interrogating the Role of the Two Distinct Fructose-Bisphosphate Aldolases of Bacillus methanolicus by Site-Directed Mutagenesis of Key Amino Acids and Gene Repression by CRISPR Interference
Front Microbiol, 12, 2021
|
|
6OPG
 
 | | phosphorylated ERK2 with AMP-PNP | | Descriptor: | MAGNESIUM ION, Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Vigers, G.P, Smith, D. | | Deposit date: | 2019-04-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Activation loop dynamics are controlled by conformation-selective inhibitors of ERK2.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6ODI
 
 | | Structure of CagY from a cryo-EM reconstruction of a T4SS | | Descriptor: | Type IV secretion system apparatus protein CagY | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-26 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
6TX6
 
 | |
7P4R
 
 | | Ultra High Resolution X-ray Structure of Orthorhombic Bovine Pancreatic Ribonuclease at 100K | | Descriptor: | ETHANOL, Ribonuclease pancreatic, SULFATE ION | | Authors: | Lisgarten, D.R, Palmer, R.A, Cooper, J.B, Naylor, C.E, Howlin, B.J, Lisgarten, J.N, Najmudin, S, Lobley, C.M.C. | | Deposit date: | 2021-07-12 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Ultra-high resolution X-ray structure of orthorhombic bovine pancreatic Ribonuclease A at 100K.
BMC Chem, 17, 2023
|
|
6TX5
 
 | |
8DXL
 
 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 4-iodopyrazole at multiple sites | | Descriptor: | 1,2-ETHANEDIOL, 4-IODOPYRAZOLE, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
8DX2
 
 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 4-amino-3-bromopyridine at multiple sites | | Descriptor: | 1,2-ETHANEDIOL, 3-bromopyridin-4-amine, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
4XAK
 
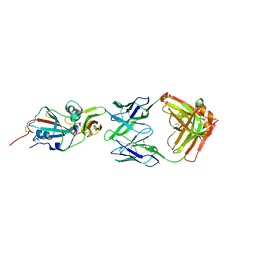 | | Crystal structure of potent neutralizing antibody m336 in complex with MERS Co-V RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Heavy chain of neutralizing antibody m336, ... | | Authors: | Zhou, T, Dimtrov, D.S, Ying, T. | | Deposit date: | 2014-12-15 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Junctional and allele-specific residues are critical for MERS-CoV neutralization by an exceptionally potent germline-like antibody.
Nat Commun, 6, 2015
|
|
8DX3
 
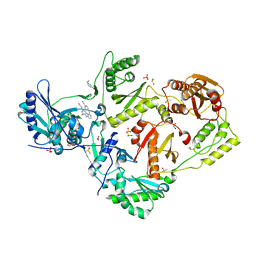 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 3-bromobenzylamine in the thumb subdomain | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-bromophenyl)methanamine, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
6EAC
 
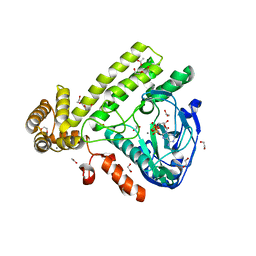 | | Pseudomonas syringae SelO | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Tomchick, D.R, Tagliabracci, V.S, Sreelatha, A. | | Deposit date: | 2018-08-02 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.269 Å) | | Cite: | Protein AMPylation by an Evolutionarily Conserved Pseudokinase.
Cell, 175, 2018
|
|
8DXE
 
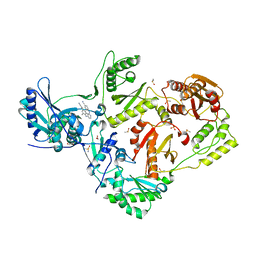 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 2-amino-6-fluorobenzonitrile at the NNRTI adjacent site | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-6-fluorobenzonitrile, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
8DXJ
 
 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 1-N-methyl-4-(trifluoromethyl)benzene-1,2-diamine at the NNRTI adjacent site | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
6OLX
 
 | | Hsp90-alpha S52A bound to PU-11-trans | | Descriptor: | 9-[(2E)-but-2-en-1-yl]-8-[(3,4,5-trimethoxyphenyl)methyl]-9H-purin-6-amine, Heat shock protein HSP 90-alpha | | Authors: | Gewirth, D.T, Huck, J.D. | | Deposit date: | 2019-04-17 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.43760586 Å) | | Cite: | Structures of Hsp90 alpha and Hsp90 beta bound to a purine-scaffold inhibitor reveal an exploitable residue for drug selectivity.
Proteins, 87, 2019
|
|
7SAQ
 
 | | Cryo-EM structure of TMEM106B fibrils extracted from a FTLD-TDP patient, polymorph 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Cao, Q, Jiang, Y, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2021-09-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Amyloid fibrils in FTLD-TDP are composed of TMEM106B and not TDP-43.
Nature, 605, 2022
|
|
6U6X
 
 | | Human SAMHD1 bound to deoxyribo(C*G*C*C*T)-oligonucleotide | | Descriptor: | DNA SC-GS-SC-SC-DT, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ZINC ION | | Authors: | Taylor, A.B, Bhattacharya, A, Wang, Z, Ivanov, D.N. | | Deposit date: | 2019-08-30 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Nucleic acid binding by SAMHD1 contributes to the antiretroviral activity and is enhanced by the GpsN modification.
Nat Commun, 12, 2021
|
|
7SAR
 
 | | Cryo-EM structure of TMEM106B fibrils extracted from a FTLD-TDP patient, polymorph 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Cao, Q, Jiang, Y, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2021-09-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Amyloid fibrils in FTLD-TDP are composed of TMEM106B and not TDP-43.
Nature, 605, 2022
|
|
4Y2D
 
 | | Crystal structure of the mCD1d/7DW8-5/iNKTCR ternary complex | | Descriptor: | 11-(4-fluorophenyl)-N-[(2S,3S,4R)-1-(alpha-D-galactopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]undecanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M, Yu, E.D. | | Deposit date: | 2015-02-09 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural modifications of alphaGalCer in both lipid and carbohydrate moiety influence activation of murine and human iNKT cells
To Be Published
|
|
6O4Y
 
 | | Structure of HLA-A2:01 with peptide MM91 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, MHC class I antigen, ... | | Authors: | Ying, G, Bitra, A, Zajonc, D.M. | | Deposit date: | 2019-03-01 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Anin silico-in vitroPipeline Identifying an HLA-A*02:01+KRAS G12V+Spliced Epitope Candidate for a Broad Tumor-Immune Response in Cancer Patients.
Front Immunol, 10, 2019
|
|
7ON7
 
 | |
7ON6
 
 | | Crystal structure of the computationally designed SAKe6AE protein | | Descriptor: | SAKe6AE, SULFATE ION | | Authors: | Wouters, S.M.L, Noguchi, H, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-05-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
8E1D
 
 | |
7OPV
 
 | | Crystal structure of the computationally designed SAKe6BE-3HH protein, alternative packing | | Descriptor: | SAKe6BE-3HH | | Authors: | Wouters, S.M.L, Noguchi, H, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-06-01 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
