2OLA
 
 | | Crystal structure of O-succinylbenzoic acid synthetase from Staphylococcus aureus, cubic crystal form | | Descriptor: | O-succinylbenzoic acid synthetase | | Authors: | Patskovsky, Y, Sauder, J.M, Ozyurt, S, Wasserman, S.R, Smith, D, Dickey, M, Maletic, M, Reyes, C, Gheyi, T, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-18 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6QN9
 
 | | Structure of bovine anti-RSV Fab B4 | | Descriptor: | GLYCEROL, Heavy chain, SULFATE ION, ... | | Authors: | Ren, J, Nettleship, J.E, Harris, G, Mwangi, W, Rhaman, N, Grant, C, Kotecha, A, Fry, E, Charleston, B, Stuart, D.I, Hammond, J, Owens, R.J. | | Deposit date: | 2019-02-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The role of the light chain in the structure and binding activity of two cattle antibodies that neutralize bovine respiratory syncytial virus.
Mol.Immunol., 112, 2019
|
|
6ZHB
 
 | | 3D electron diffraction structure of bovine insulin | | Descriptor: | Insulin, ZINC ION | | Authors: | Blum, T, Housset, D, Clabbers, M.T.B, van Genderen, E, Bacia-Verloop, M, Zander, U, McCarthy, A.A, Schoehn, G, Ling, W.L, Abrahams, J.P. | | Deposit date: | 2020-06-22 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.25 Å) | | Cite: | Statistically correcting dynamical electron scattering improves the refinement of protein nanocrystals, including charge refinement of coordinated metals.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6EKK
 
 | | Crystal structure of GEF domain of DENND 1A in complex with Rab GTPase Rab35-GDP bound state. | | Descriptor: | 1,2-ETHANEDIOL, DENN domain-containing protein 1A, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Srikannathasan, V, Szykowska, A, Tallant, C, Strain-Damerell, C, Kopec, J, Kupinska, K, Mukhopadhyay, S, Gavin, M, Wang, D, Chalk, R, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Huber, K. | | Deposit date: | 2017-09-26 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of DENND1A-RAB35 complex with GDP bound state.
To be published
|
|
2DJX
 
 | | Crystal structure of native Trypanosoma cruzi dihydroorotate dehydrogenase | | Descriptor: | COBALT HEXAMMINE(III), Dihydroorotate Dehydrogenase, FLAVIN MONONUCLEOTIDE | | Authors: | Inaoka, D.K, Shimizu, H, Sakamoto, K, Shiba, T, Kurisu, G, Nara, T, Aoki, T, Harada, S, Kita, K. | | Deposit date: | 2006-04-05 | | Release date: | 2007-06-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of native Trypanosoma cruzi dihydroorotate dehydrogenase
To be Published
|
|
6ELN
 
 | | Estimation of relative drug-target residence times by random acceleration molecular dynamics simulation | | Descriptor: | 4-[4-(4-methoxyphenyl)-5-methyl-1H-pyrazol-3-yl]benzene-1,3-diol, Heat shock protein HSP 90-alpha, SULFATE ION | | Authors: | Musil, D, Lehmann, M, Eggenweiler, H.-M. | | Deposit date: | 2017-09-29 | | Release date: | 2018-05-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Estimation of Drug-Target Residence Times by tau-Random Acceleration Molecular Dynamics Simulations.
J Chem Theory Comput, 14, 2018
|
|
6QLS
 
 | | Galectin-3C in complex with fluoroaryltriazole monothiogalactoside derivative 6 | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,5-bis(oxidanyl)-4-(4-phenyl-1,2,3-triazol-1-yl)oxan-2-yl]sulfanyl-oxane-3,4,5-triol, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-02-01 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.047 Å) | | Cite: | Structure and Energetics of Ligand-Fluorine Interactions with Galectin-3 Backbone and Side-Chain Amides: Insight into Solvation Effects and Multipolar Interactions.
Chemmedchem, 14, 2019
|
|
6ZFM
 
 | | Structure of alpha-Cobratoxin with a peptide inhibitor | | Descriptor: | 3-[2-[2-[2-[2-[2-(2-azanylethoxy)ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]propan-1-ol, Alpha-cobratoxin, PENTAETHYLENE GLYCOL, ... | | Authors: | Kiontke, S, Kummel, D. | | Deposit date: | 2020-06-17 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Peptide Inhibitors of the alpha-Cobratoxin-Nicotinic Acetylcholine Receptor Interaction.
J.Med.Chem., 63, 2020
|
|
8BIK
 
 | | Crystal structure of human AMPK heterotrimer in complex with allosteric activator C455 | | Descriptor: | (3~{R},3~{a}~{R},6~{R},6~{a}~{R})-6-[[6-chloranyl-5-[4-[4-[[dimethyl(oxidanyl)-$l^{4}-sulfanyl]amino]phenyl]phenyl]-3~{H}-imidazo[4,5-b]pyridin-2-yl]oxy]-2,3,3~{a},5,6,6~{a}-hexahydrofuro[3,2-b]furan-3-ol, 5'-AMP-activated protein kinase catalytic subunit alpha-2, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Schimpl, M, Mather, K.M, Boland, M.L, Rivers, E.L, Srivastava, A, Hemsley, P, Robinson, J, Wan, P.T, Hansen, J, Read, J.A, Trevaskis, J.L, Smith, D.M. | | Deposit date: | 2022-11-02 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Direct beta 1/ beta 2 AMPK activation reduces liver steatosis but not fibrosis in a mouse model of non-alcoholic steatohepatitis
Biorxiv, 2024
|
|
4LQV
 
 | | Thrirty minutes iron loaded frog M ferritin | | Descriptor: | CHLORIDE ION, FE (II) ION, Ferritin, ... | | Authors: | Mangani, S, Di Pisa, F, Pozzi, C, Turano, P, Lalli, D. | | Deposit date: | 2013-07-19 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Time-lapse anomalous X-ray diffraction shows how Fe(2+) substrate ions move through ferritin protein nanocages to oxidoreductase sites.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6ZI8
 
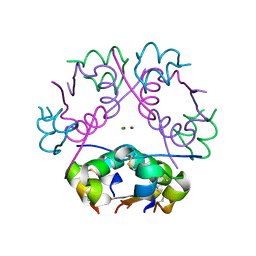 | | X-ray diffraction structure of bovine insulin at 2.3 A resolution | | Descriptor: | CHLORIDE ION, Insulin, ZINC ION | | Authors: | Housset, D, Ling, W.L, Bacia-Verloop, M, Zander, U, McCarthy, A.A, Schoehn, G. | | Deposit date: | 2020-06-25 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Statistically correcting dynamical electron scattering improves the refinement of protein nanocrystals, including charge refinement of coordinated metals.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6QSD
 
 | | Crystal structure of Pizza6S | | Descriptor: | Pizza6S, SULFATE ION | | Authors: | Noguchi, H, Vandebroek, L, Kamata, K, Tame, J.R.H, Van Meervelt, L, Parac-Vogt, T.N, Voet, A.R.D. | | Deposit date: | 2019-02-20 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Hybrid assemblies of a symmetric designer protein and polyoxometalates with matching symmetry.
Chem.Commun.(Camb.), 56, 2020
|
|
6QSH
 
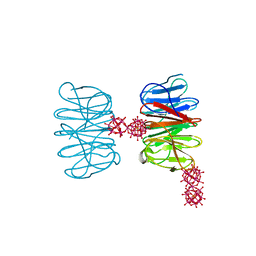 | | Crystal structure of the hybrid bioinorganic complex of Pizza6S linked by the 1:2 Ce-substituted Keggin | | Descriptor: | 1:2 Ce-substituted Keggin, Pizza6S | | Authors: | Noguchi, H, Vandebroek, L, Kamata, K, Tame, J.R.H, Van Meervelt, L, Parac-Vogt, T.N, Voet, A.R.D. | | Deposit date: | 2019-02-20 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Hybrid assemblies of a symmetric designer protein and polyoxometalates with matching symmetry.
Chem.Commun.(Camb.), 56, 2020
|
|
6QU9
 
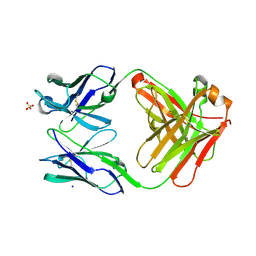 | | Fab fragment of an antibody that inhibits polymerisation of alpha-1-antitrypsin | | Descriptor: | FAB 4B12 heavy chain, FAB 4B12 light chain, GLYCEROL, ... | | Authors: | Jagger, A.M, Heyer-Chauhan, N, Lomas, D.A, Irving, J.A. | | Deposit date: | 2019-02-26 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for Z alpha 1 -antitrypsin polymerization in the liver.
Sci Adv, 6, 2020
|
|
5XNJ
 
 | |
6YVY
 
 | |
1U9G
 
 | | Heterocyclic Peptide Backbone Modification in GCN4-pLI Based Coiled Coils: Replacement of K(8)L(9) | | Descriptor: | General control protein GCN4, SULFATE ION | | Authors: | Horne, W.S, Yadav, M.K, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2004-08-09 | | Release date: | 2004-11-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Heterocyclic peptide backbone modifications in an alpha-helical coiled coil.
J.Am.Chem.Soc., 126, 2004
|
|
6YVS
 
 | | FOCAL ADHESION KINASE CATALYTIC DOMAIN IN COMPLEX WITH 5-{4-[(Pyridin-3-ylmethyl)-amino]-5-trifluoromethyl-pyrimidin-2-ylamino}-1,3-dihydro-indol-2-one | | Descriptor: | 5-[[4-(pyridin-3-ylmethylamino)-5-(trifluoromethyl)pyrimidin-2-yl]amino]-1,3-dihydroindol-2-one, Focal adhesion kinase 1, SULFATE ION | | Authors: | Musil, D, Heinrich, T, Amaral, M. | | Deposit date: | 2020-04-28 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure-kinetic relationship reveals the mechanism of selectivity of FAK inhibitors over PYK2.
Cell Chem Biol, 28, 2021
|
|
6YXV
 
 | |
6QZ6
 
 | | Structure of Mcl-1 in complex with compound 8b | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
7NYD
 
 | | cryoEM structure of 2C9-sMAC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Menny, A, Couves, E.C, Bubeck, D. | | Deposit date: | 2021-03-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural basis of soluble membrane attack complex packaging for clearance.
Nat Commun, 12, 2021
|
|
4DDF
 
 | | Computationally Designed Self-assembling Octahedral Cage protein, O333, Crystallized in space group P4 | | Descriptor: | CHLORIDE ION, Propanediol utilization polyhedral body protein PduT, SULFATE ION | | Authors: | Sawaya, M.R, King, N.P, Sheffler, W, Baker, D, Yeates, T.O. | | Deposit date: | 2012-01-18 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Computational design of self-assembling protein nanomaterials with atomic level accuracy.
Science, 336, 2012
|
|
6QYP
 
 | | Structure of Mcl-1 in complex with compound 13 | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-5-(4-methylpiperazin-1-yl)-4-oxidanyl-phenyl]-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QZ5
 
 | | Structure of Mcl-1 in complex with compound 8a | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QZ4
 
 | | Structure of MHETase from Ideonella sakaiensis | | Descriptor: | CALCIUM ION, Mono(2-hydroxyethyl) terephthalate hydrolase, SULFATE ION | | Authors: | Allen, M.D, Johnson, C.W, Knott, B.C, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2019-03-11 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization and engineering of a two-enzyme system for plastics depolymerization.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
