7L7I
 
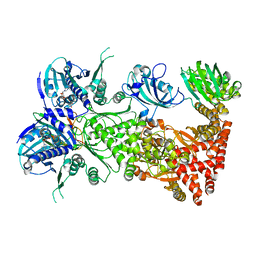 | | Cryo-EM structure of Hsp90:FKBP51:p23 closed-state complex | | Descriptor: | Heat shock protein HSP 90-alpha, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Peptidyl-prolyl cis-trans isomerase FKBP5, ... | | Authors: | Lee, K, Thwin, A.C, Tse, E, Gates, S.N, Southworth, D.R. | | Deposit date: | 2020-12-28 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structure of an Hsp90-immunophilin complex reveals cochaperone recognition of the client maturation state.
Mol.Cell, 81, 2021
|
|
2MGO
 
 | | NMR solution structure of oxytocin | | Descriptor: | Oxytocin | | Authors: | Harvey, P.J, Craik, D.J. | | Deposit date: | 2013-11-01 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Oxytocic plant cyclotides as templates for peptide G protein-coupled receptor ligand design.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
177D
 
 | |
2LZG
 
 | | NMR Structure of Mdm2 (6-125) with Pip-1 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, [(3R,5R,6S)-5-(3-chlorophenyl)-6-(4-chlorophenyl)-1-(cyclopropylmethyl)-2-oxopiperidin-3-yl]acetic acid | | Authors: | Michelsen, K.B, Jordan, J.B, Lewis, J, Long, A.M, Yang, E, Rew, Y, Zhou, J, Yakowec, P, Schnier, P.D, Huang, X, Poppe, L. | | Deposit date: | 2012-10-02 | | Release date: | 2012-11-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Ordering of the N-Terminus of Human MDM2 by Small Molecule Inhibitors.
J.Am.Chem.Soc., 134, 2012
|
|
2LXA
 
 | |
2M1M
 
 | | Solution structure of the PsIAA4 oligomerization domain reveals interaction modes for transcription factors in early auxin response | | Descriptor: | Auxin-induced protein IAA4 | | Authors: | Kovermann, M, Dinesh, D.C, Gopalswamy, M, Abel, S, Balbach, J. | | Deposit date: | 2012-12-03 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PsIAA4 oligomerization domain reveals interaction modes for transcription factors in early auxin response.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6V3C
 
 | |
2M3T
 
 | |
2FCI
 
 | | Structural basis for the requirement of two phosphotyrosines in signaling mediated by Syk tyrosine kinase | | Descriptor: | C-termainl SH2 domain from phospholipase C-gamma-1 comprising residues 663-759, Doubly phosphorylated peptide derived from Syk kinase comprising residues 338-350 | | Authors: | Groesch, T.D, Zhou, F, Mattila, S, Geahlen, R.L, Post, C.B. | | Deposit date: | 2005-12-12 | | Release date: | 2006-01-31 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the requirement of two phosphotyrosine residues in signaling mediated by syk tyrosine kinase
J.Mol.Biol., 356, 2006
|
|
8WFE
 
 | | The Crystal Structure of PPARg from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor gamma | | Authors: | Wang, F, Cheng, W, Lv, Z, Guo, S, Lin, D. | | Deposit date: | 2023-09-19 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of PPARg from Biortus.
To Be Published
|
|
5FO3
 
 | | ZapC cell division regulator from E. coli | | Descriptor: | CELL DIVISION PROTEIN ZAPC | | Authors: | Ortiz, C, Kureisaite-Ciziene, D, Schmitz, F, Vicente, M, Lowe, J. | | Deposit date: | 2015-11-18 | | Release date: | 2015-11-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Z-Ring Associated Cell Division Protein Zapc from Escherichia Coli.
FEBS Lett., 589, 2015
|
|
5I4H
 
 | |
8QYO
 
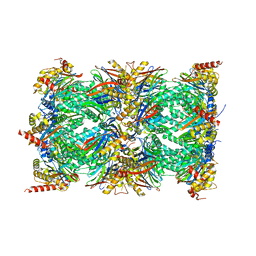 | | Human proteasome 20S core particle | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Schulman, B.A, Hanna, J.W, Harper, J.W, Adolf, F, Du, J, Rawson, S.D, Walsh Jr, R.M, Goodall, E.A. | | Deposit date: | 2023-10-26 | | Release date: | 2024-02-21 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Visualizing chaperone-mediated multistep assembly of the human 20S proteasome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
2M8J
 
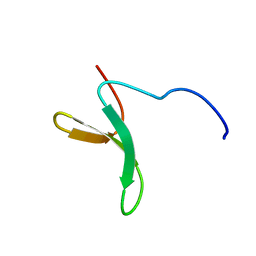 | | Structure of Pin1 WW domain phospho-mimic S16E | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Luh, L.M, Kirchner, D.K, Loehr, F, Haensel, R, Doetsch, V. | | Deposit date: | 2013-05-22 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular crowding drives active Pin1 into nonspecific complexes with endogenous proteins prior to substrate recognition.
J.Am.Chem.Soc., 135, 2013
|
|
3EFL
 
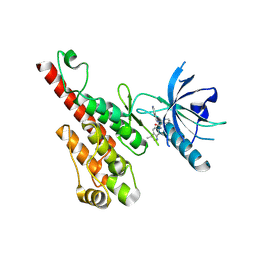 | | Crystal structure of the VEGFR2 kinase domain in complex with motesanib | | Descriptor: | N-(3,3-dimethyl-2,3-dihydro-1H-indol-6-yl)-2-[(pyridin-4-ylmethyl)amino]pyridine-3-carboxamide, Vascular endothelial growth factor receptor 2 | | Authors: | Kim, J.L, Whittington, D.A, Long, A.M, Rose, P, Gu, Y, Zhao, H. | | Deposit date: | 2008-09-09 | | Release date: | 2009-09-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Motesanib
To be Published
|
|
2JM4
 
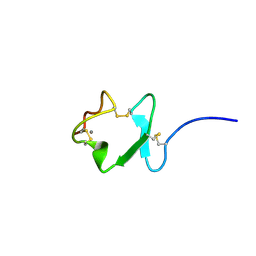 | |
2JST
 
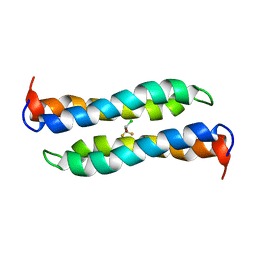 | | Four-Alpha-Helix Bundle with Designed Anesthetic Binding Pockets II: Halothane Effects on Structure and Dynamics | | Descriptor: | 2-BROMO-2-CHLORO-1,1,1-TRIFLUOROETHANE, Four-Alpha-Helix Bundle | | Authors: | Cui, T, Bondarenko, V, Ma, D, Canlas, C, Brandon, N.R, Johansson, J.S, Tang, P, Xu, Y. | | Deposit date: | 2007-07-12 | | Release date: | 2008-05-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Four-alpha-helix bundle with designed anesthetic binding pockets. Part II: halothane effects on structure and dynamics
Biophys.J., 94, 2008
|
|
7AHI
 
 | | Substrate-engaged type 3 secretion system needle complex from Salmonella enterica typhimurium - SpaR state 2 | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, LAURYL DIMETHYLAMINE-N-OXIDE, Lipoprotein PrgK, ... | | Authors: | Fahrenkamp, D, Goessweiner-Mohr, N, Miletic, S, Wald, J, Marlovits, T. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Substrate-engaged type III secretion system structures reveal gating mechanism for unfolded protein translocation
Nat Commun, 12, 2021
|
|
2JYT
 
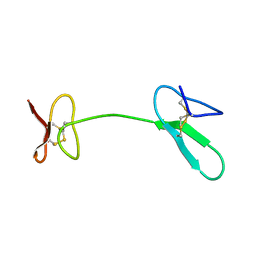 | | Human Granulin C, isomer 1 | | Descriptor: | Granulin-5 | | Authors: | Tolkatchev, D, Wang, P, Chen, Z, Xu, P, Ni, F. | | Deposit date: | 2007-12-19 | | Release date: | 2008-04-22 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure dissection of human progranulin identifies well-folded granulin/epithelin modules with unique functional activities.
Protein Sci., 17, 2008
|
|
8CH7
 
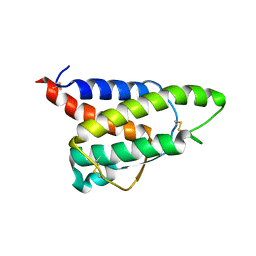 | | RDC-refined Interleukin-4 (wild type) pH 5.6 | | Descriptor: | Interleukin-4 | | Authors: | Vaz, D.C, Rodrigues, J.R, Loureiro-Ferreira, N, Mueller, T, Sebald, W, Redfield, C, Brito, R.M.M. | | Deposit date: | 2023-02-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Lessons on protein structure from interleukin-4: All disulfides are not created equal.
Proteins, 92, 2024
|
|
2JZP
 
 | | NMR solution structure of Kx5Q ProtL mutant | | Descriptor: | Protein L | | Authors: | Lopez-Mendez, N, Tadeo, X, Castano, D, Pons, M, Millet Aguilar-Galindo, O. | | Deposit date: | 2008-01-11 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Protein stabilization and the hofmeister effect: the role of hydrophobic solvation
Biophys.J., 97, 2009
|
|
8CGF
 
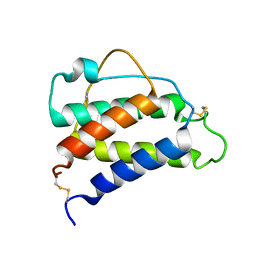 | | Interleukin-4 (wild type) pH 2.4 | | Descriptor: | Interleukin-4 | | Authors: | Vaz, D.C, Rodrigues, J.R, Loureiro-Ferreira, N, Mueller, T, Sebald, W, Redfield, C, Brito, R.M.M. | | Deposit date: | 2023-02-04 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Lessons on protein structure from interleukin-4: All disulfides are not created equal.
Proteins, 92, 2024
|
|
7ME2
 
 | | YfeA oligomer crystal 2, form 2 | | Descriptor: | FE (III) ION, MANGANESE (II) ION, Periplasmic chelated iron-binding protein YfeA, ... | | Authors: | Radka, C.D, Aller, S.G. | | Deposit date: | 2021-04-06 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Site 2 of the Yersinia pestis substrate-binding protein YfeA is a dynamic surface metal-binding site
Acta Crystallogr.,Sect.F, 77, 2021
|
|
5IHW
 
 | | The crystal structure of SdrE from staphylococcus aureus | | Descriptor: | Serine-aspartate repeat-containing protein E | | Authors: | Zhang, S, Wei, J, Wu, S, Zhang, X, Luo, M, Wang, D. | | Deposit date: | 2016-02-29 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The crystal structure of SdrE from staphylococcus aureus
To Be Published
|
|
7ME3
 
 | | YfeA oligomer crystal 3, form 2 | | Descriptor: | FE (III) ION, MANGANESE (II) ION, Periplasmic chelated iron-binding protein YfeA, ... | | Authors: | Radka, C.D, Aller, S.G. | | Deposit date: | 2021-04-06 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Site 2 of the Yersinia pestis substrate-binding protein YfeA is a dynamic surface metal-binding site
Acta Crystallogr.,Sect.F, 77, 2021
|
|
